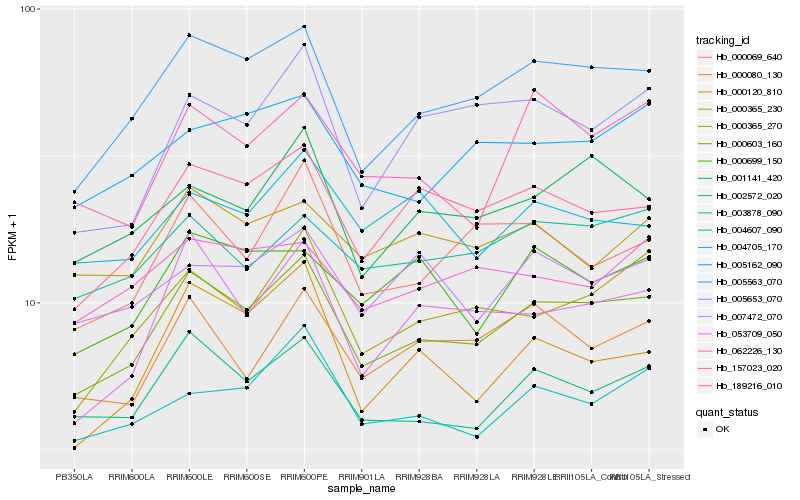
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000603_160 |
0.0 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 2 |
Hb_005563_070 |
0.0440439672 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 3 |
Hb_002572_020 |
0.0611701141 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 4 |
Hb_189216_010 |
0.0644381644 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000365_270 |
0.0703977727 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 6 |
Hb_001141_420 |
0.0726451762 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004607_090 |
0.0749872938 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 8 |
Hb_053709_050 |
0.0756350399 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_000069_640 |
0.0787465451 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 10 |
Hb_003878_090 |
0.0787789767 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 11 |
Hb_000080_130 |
0.0788820389 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 12 |
Hb_005162_090 |
0.0801971951 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 13 |
Hb_157023_020 |
0.0832898856 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000699_150 |
0.0840195826 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 15 |
Hb_005653_070 |
0.0848346899 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 16 |
Hb_000120_810 |
0.0852251842 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 17 |
Hb_007472_070 |
0.0855481453 |
- |
- |
cir, putative [Ricinus communis] |
| 18 |
Hb_000365_230 |
0.0861076455 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 19 |
Hb_062226_130 |
0.0874989761 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 20 |
Hb_004705_170 |
0.0878042691 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |