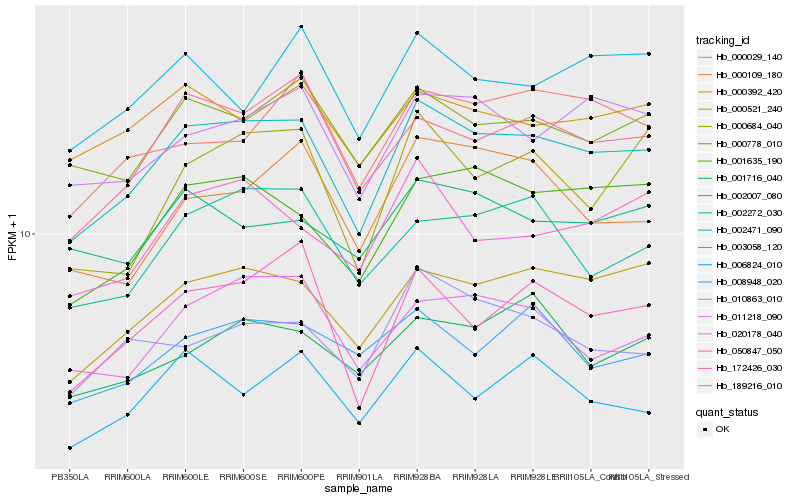
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 2 |
Hb_000521_240 |
0.0605746885 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 3 |
Hb_189216_010 |
0.0681223725 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001635_190 |
0.0689806667 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000778_010 |
0.0727368327 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 6 |
Hb_011218_090 |
0.0754625194 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 7 |
Hb_020178_040 |
0.0785083414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_172426_030 |
0.0787200781 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 9 |
Hb_002272_030 |
0.0800639601 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 10 |
Hb_000392_420 |
0.0811880847 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 11 |
Hb_050847_050 |
0.0818858353 |
- |
- |
PREDICTED: uncharacterized protein LOC105649140 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000029_140 |
0.0849007987 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 13 |
Hb_010863_010 |
0.0857614321 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_000684_040 |
0.0862798216 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 15 |
Hb_002007_080 |
0.0867075903 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_001716_040 |
0.0876429492 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 17 |
Hb_008948_020 |
0.0878467211 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 18 |
Hb_006824_010 |
0.0883378718 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 19 |
Hb_003058_120 |
0.0887149873 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 20 |
Hb_000109_180 |
0.0894629955 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |