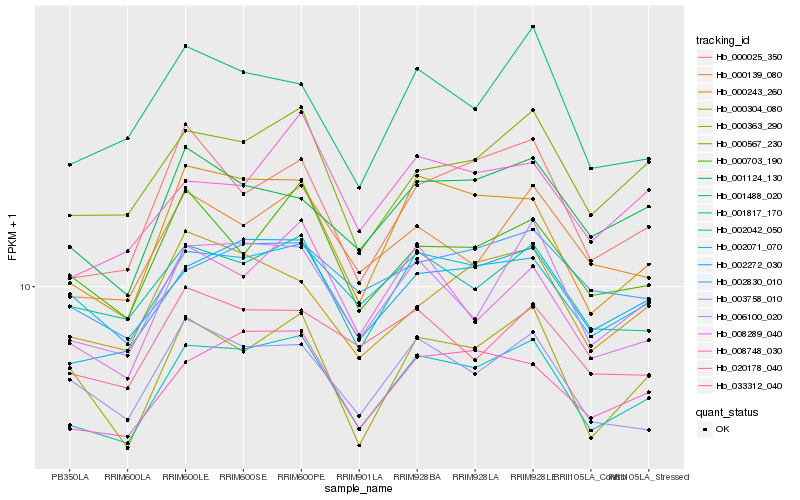
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002272_030 |
0.0421181025 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 3 |
Hb_020178_040 |
0.0525924528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000243_260 |
0.0569452305 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006100_020 |
0.0572473459 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 6 |
Hb_000567_230 |
0.058187878 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 7 |
Hb_001817_170 |
0.0583150063 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 8 |
Hb_002071_070 |
0.0584942107 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000139_080 |
0.067028824 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001124_130 |
0.0684926622 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 11 |
Hb_000363_290 |
0.0697790296 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 12 |
Hb_000025_350 |
0.0704081599 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000703_190 |
0.072097103 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003758_010 |
0.0722640016 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008289_040 |
0.0731684937 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 16 |
Hb_008748_030 |
0.0735470629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000304_080 |
0.0738054403 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_033312_040 |
0.074805115 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002830_010 |
0.0753856387 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_001488_020 |
0.0758234646 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |