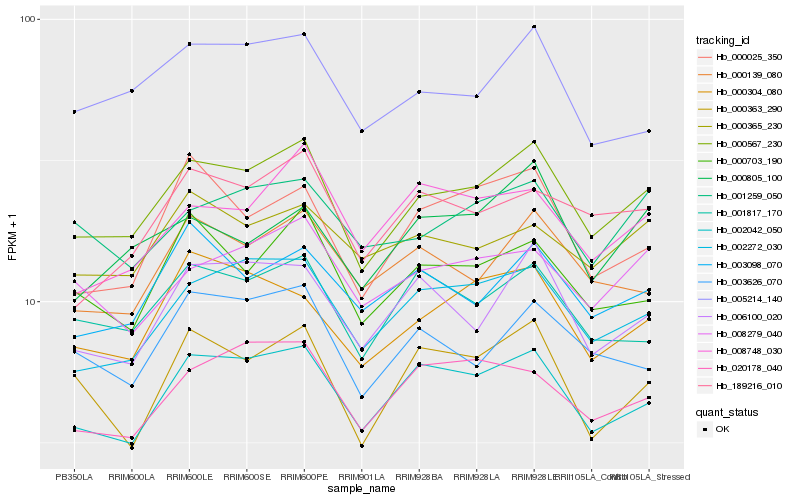
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_230 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 2 |
Hb_002042_050 |
0.058187878 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002272_030 |
0.0620159701 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 4 |
Hb_001817_170 |
0.0727422075 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 5 |
Hb_000703_190 |
0.0739319234 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006100_020 |
0.074666011 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 7 |
Hb_000139_080 |
0.0755009622 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_008279_040 |
0.0760934785 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_005214_140 |
0.0764415923 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 10 |
Hb_000304_080 |
0.0769840517 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001259_050 |
0.0776368168 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_003098_070 |
0.0781959398 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 13 |
Hb_003626_070 |
0.0783676689 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 14 |
Hb_189216_010 |
0.0794128092 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 15 |
Hb_020178_040 |
0.0795899539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000805_100 |
0.0809266386 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000025_350 |
0.0816286063 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000365_230 |
0.0816820937 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 19 |
Hb_000363_290 |
0.0819064015 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 20 |
Hb_008748_030 |
0.0827005112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |