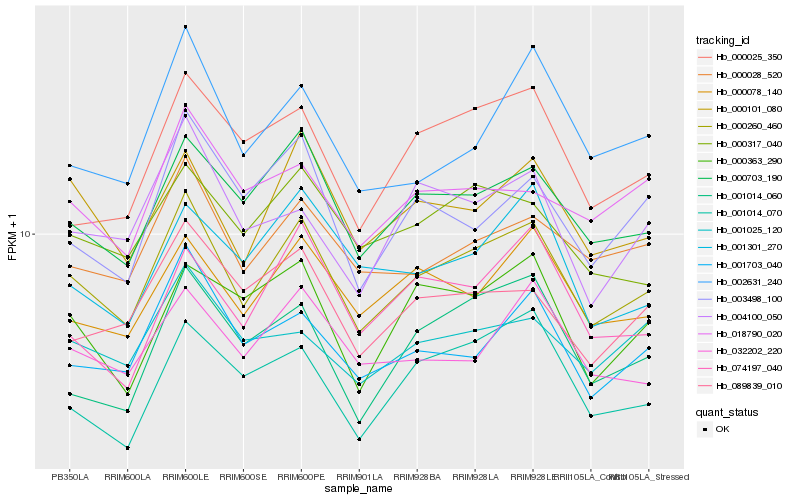
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_460 |
0.0 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000703_190 |
0.0614051854 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000101_080 |
0.069905719 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 4 |
Hb_000028_520 |
0.0752578532 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 5 |
Hb_000078_140 |
0.0764525072 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_000317_040 |
0.0793548916 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_001703_040 |
0.0803555889 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 8 |
Hb_032202_220 |
0.0814262192 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000025_350 |
0.0845079086 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_089839_010 |
0.0858256834 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 11 |
Hb_002631_240 |
0.0879028939 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 12 |
Hb_000363_290 |
0.0879318567 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 13 |
Hb_001014_060 |
0.0879757041 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 14 |
Hb_001014_070 |
0.0883328074 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 15 |
Hb_074197_040 |
0.0891521889 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 16 |
Hb_001025_120 |
0.0898738592 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 17 |
Hb_001301_270 |
0.0901076122 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 18 |
Hb_004100_050 |
0.0901255297 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |
| 19 |
Hb_003498_100 |
0.0901665506 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 20 |
Hb_018790_020 |
0.0909790801 |
- |
- |
protein kinase, putative [Ricinus communis] |