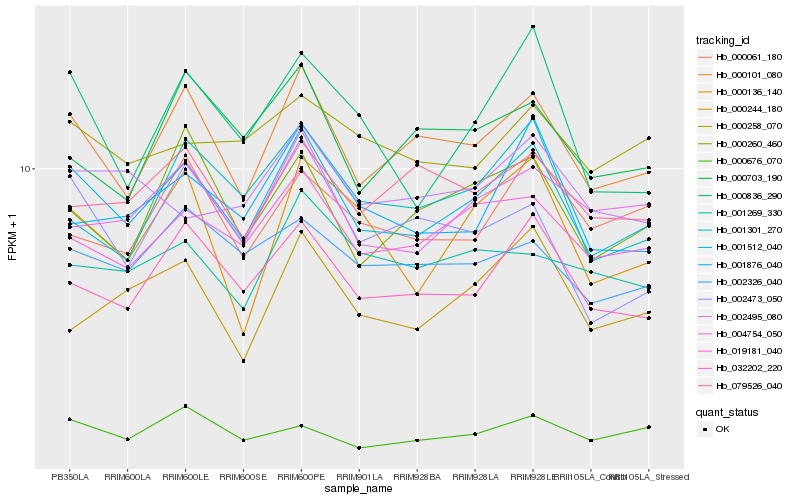
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001512_040 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 2 |
Hb_000101_080 |
0.0594410173 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 3 |
Hb_032202_220 |
0.0742736194 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_019181_040 |
0.077129579 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 5 |
Hb_004754_050 |
0.0878809104 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 6 |
Hb_002326_040 |
0.091312674 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 7 |
Hb_000836_290 |
0.0913395083 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001876_040 |
0.0913732061 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 9 |
Hb_000260_460 |
0.0927537792 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001301_270 |
0.0962516293 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 11 |
Hb_000676_070 |
0.0973041608 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 12 |
Hb_000136_140 |
0.0974331522 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 13 |
Hb_002473_050 |
0.0986622952 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_079526_040 |
0.0991397295 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_000258_070 |
0.1000821884 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 16 |
Hb_000244_180 |
0.1004731829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000061_180 |
0.1013280134 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 18 |
Hb_002495_080 |
0.1020481613 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 19 |
Hb_001269_330 |
0.1022050226 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_000703_190 |
0.1023773086 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |