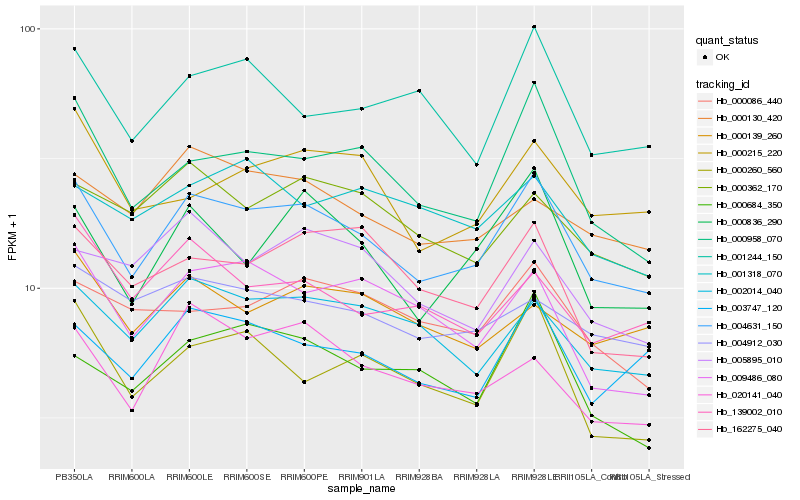
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_150 |
0.0 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000958_070 |
0.0809031579 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 3 |
Hb_003747_120 |
0.0846492864 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000836_290 |
0.0856531796 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002014_040 |
0.0858451236 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 6 |
Hb_000362_170 |
0.0961059104 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 7 |
Hb_001244_150 |
0.0963019 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 8 |
Hb_162275_040 |
0.0973156365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_009486_080 |
0.0974448194 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 10 |
Hb_000215_220 |
0.0975795108 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000130_420 |
0.0975881414 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_000086_440 |
0.097719991 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 13 |
Hb_020141_040 |
0.0979400084 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004912_030 |
0.0989791839 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 15 |
Hb_000260_560 |
0.0993369295 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000684_350 |
0.0994271264 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005895_010 |
0.102453298 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 18 |
Hb_139002_010 |
0.1033190904 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000139_260 |
0.1037907439 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 20 |
Hb_001318_070 |
0.1061965602 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |