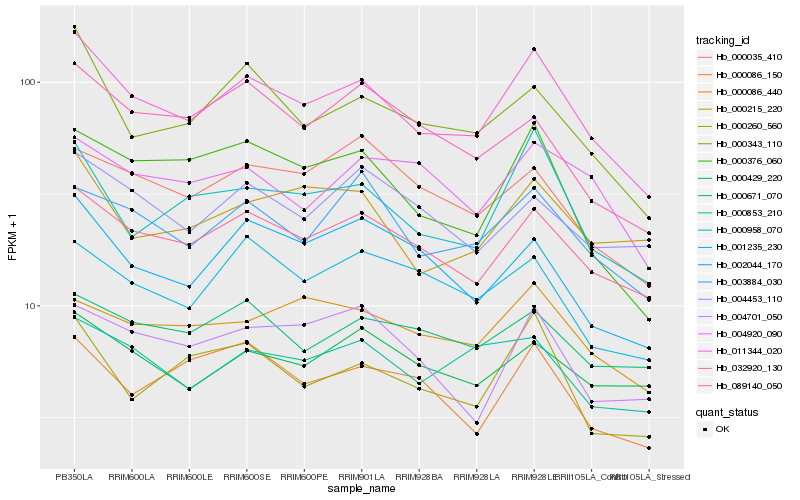
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011344_020 |
0.0 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 2 |
Hb_089140_050 |
0.0642279676 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 3 |
Hb_000958_070 |
0.0786881126 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 4 |
Hb_000343_110 |
0.0822551016 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 5 |
Hb_003884_030 |
0.0844203306 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 6 |
Hb_000376_060 |
0.090772165 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000429_220 |
0.0945827685 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000853_210 |
0.0946912132 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_560 |
0.0953740395 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002044_170 |
0.0960950321 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 11 |
Hb_000086_440 |
0.0979923009 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 12 |
Hb_000215_220 |
0.0989498094 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001235_230 |
0.100323138 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 14 |
Hb_000671_070 |
0.1008823989 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 15 |
Hb_004453_110 |
0.1011141844 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000035_410 |
0.1022568271 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 17 |
Hb_004920_090 |
0.1023996157 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_032920_130 |
0.1036722377 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 19 |
Hb_000086_150 |
0.1038198908 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004701_050 |
0.1040066199 |
- |
- |
alpha1 family protein [Populus trichocarpa] |