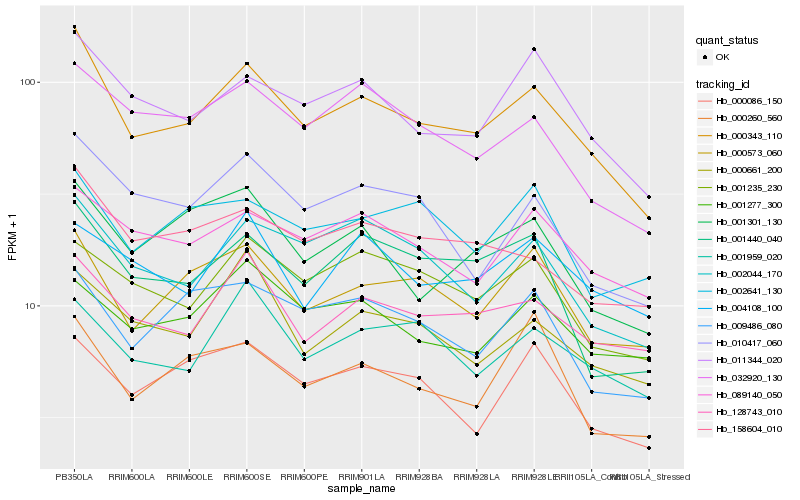
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_110 |
0.0 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 2 |
Hb_011344_020 |
0.0822551016 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 3 |
Hb_000573_060 |
0.0885773493 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 4 |
Hb_002044_170 |
0.0898968213 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 5 |
Hb_128743_010 |
0.0901915586 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_158604_010 |
0.0932528975 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 7 |
Hb_032920_130 |
0.0934071286 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 8 |
Hb_010417_060 |
0.0958016655 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 9 |
Hb_089140_050 |
0.0959227431 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 10 |
Hb_001440_040 |
0.0984798344 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 11 |
Hb_001235_230 |
0.1006036626 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_001301_130 |
0.1010409617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 13 |
Hb_000661_200 |
0.1015863117 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009486_080 |
0.1021215791 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 15 |
Hb_001959_020 |
0.1031952239 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000086_150 |
0.1058215903 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001277_300 |
0.1087534781 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 18 |
Hb_002641_130 |
0.1087926009 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 19 |
Hb_000260_560 |
0.1088716695 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004108_100 |
0.1106606302 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |