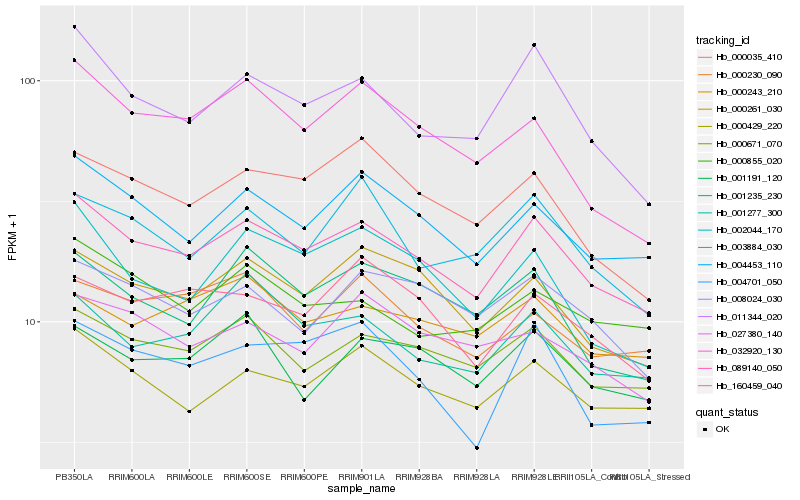
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089140_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 2 |
Hb_004453_110 |
0.0551063435 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000671_070 |
0.0571231915 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 4 |
Hb_000429_220 |
0.0606591097 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000261_030 |
0.064062165 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 6 |
Hb_011344_020 |
0.0642279676 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 7 |
Hb_000035_410 |
0.0725945564 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 8 |
Hb_001277_300 |
0.0742097043 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 9 |
Hb_008024_030 |
0.075328748 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 10 |
Hb_003884_030 |
0.0757074552 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 11 |
Hb_001235_230 |
0.0766492022 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_160459_040 |
0.0773121727 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 13 |
Hb_027380_140 |
0.0774432015 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004701_050 |
0.0785329594 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 15 |
Hb_000230_090 |
0.0787884515 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000855_020 |
0.0788020623 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 17 |
Hb_032920_130 |
0.0790721058 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 18 |
Hb_001191_120 |
0.0790831515 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000243_210 |
0.0794110762 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_002044_170 |
0.0796500268 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |