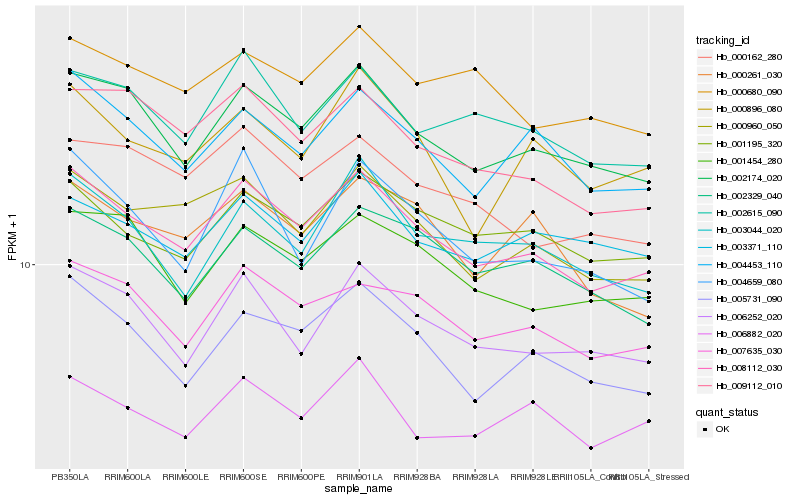
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008112_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002174_020 |
0.0451301905 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_007635_030 |
0.0520540942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000960_050 |
0.0527851351 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 5 |
Hb_004453_110 |
0.0569464147 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 6 |
Hb_005731_090 |
0.0582181896 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 7 |
Hb_006252_020 |
0.0590488028 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_009112_010 |
0.0607395423 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 9 |
Hb_001195_320 |
0.0627027687 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 10 |
Hb_001454_280 |
0.0642447373 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 11 |
Hb_002615_090 |
0.0643247096 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000162_280 |
0.0668889637 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 13 |
Hb_002329_040 |
0.0674696041 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 14 |
Hb_003044_020 |
0.0694758168 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 15 |
Hb_000896_080 |
0.0712234479 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP95 isoform X1 [Vitis vinifera] |
| 16 |
Hb_000261_030 |
0.0715658659 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 17 |
Hb_000680_090 |
0.073277787 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 18 |
Hb_006882_020 |
0.0733720208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004659_080 |
0.0734073916 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 20 |
Hb_003371_110 |
0.0734898546 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |