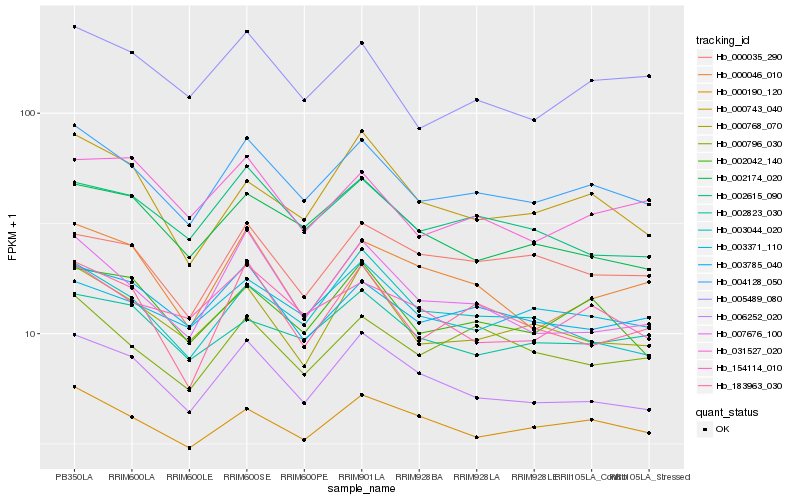
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_050 |
0.0 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_006252_020 |
0.0572648279 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003785_040 |
0.0584098253 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002042_140 |
0.0607253676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647570 [Jatropha curcas] |
| 5 |
Hb_005489_080 |
0.0626722918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000768_070 |
0.064317222 |
transcription factor |
TF Family: SNF2 |
PREDICTED: TATA-binding protein-associated factor BTAF1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000190_120 |
0.0649181888 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 8 |
Hb_000796_030 |
0.0664685133 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_154114_010 |
0.0674282693 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 10 |
Hb_002615_090 |
0.0681562987 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003371_110 |
0.0693707312 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 12 |
Hb_002174_020 |
0.0702121516 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_002823_030 |
0.0707582974 |
- |
- |
- |
| 14 |
Hb_183963_030 |
0.0721363486 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_007676_100 |
0.0721567554 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 16 |
Hb_003044_020 |
0.073680463 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 17 |
Hb_000046_010 |
0.0756502701 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 18 |
Hb_000743_040 |
0.0758598659 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000035_290 |
0.0761491506 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 20 |
Hb_031527_020 |
0.0767355599 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |