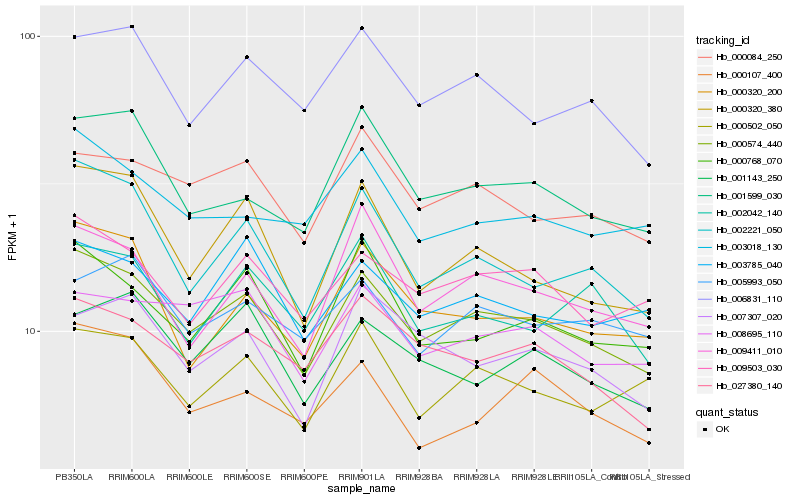
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_440 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634833 [Jatropha curcas] |
| 2 |
Hb_009503_030 |
0.0604967124 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 3 |
Hb_000084_250 |
0.0608267153 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 4 |
Hb_002221_050 |
0.0628917013 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 5 |
Hb_027380_140 |
0.0634967924 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006831_110 |
0.0643504637 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 7 |
Hb_000768_070 |
0.0649602385 |
transcription factor |
TF Family: SNF2 |
PREDICTED: TATA-binding protein-associated factor BTAF1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000502_050 |
0.0668433955 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF8, putative [Ricinus communis] |
| 9 |
Hb_003785_040 |
0.06795214 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001599_030 |
0.06838277 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 11 |
Hb_008695_110 |
0.0691627328 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009411_010 |
0.0691904226 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |
| 13 |
Hb_000320_380 |
0.0705393724 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000320_200 |
0.0713863086 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 15 |
Hb_003018_130 |
0.072317911 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 16 |
Hb_001143_250 |
0.072368763 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 17 |
Hb_005993_050 |
0.0735287392 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 18 |
Hb_007307_020 |
0.0753071089 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000107_400 |
0.0753990597 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870-like [Jatropha curcas] |
| 20 |
Hb_002042_140 |
0.0758445369 |
- |
- |
PREDICTED: uncharacterized protein LOC105647570 [Jatropha curcas] |