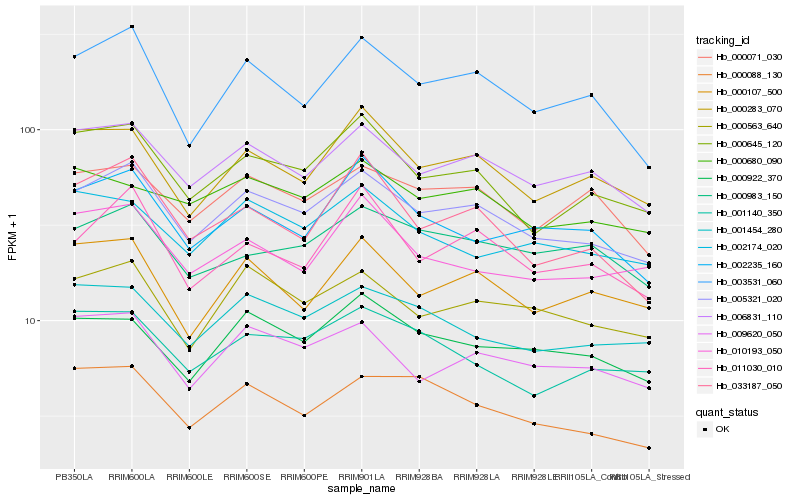
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005321_020 |
0.0 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 2 |
Hb_006831_110 |
0.0522175519 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 3 |
Hb_000563_640 |
0.0621069781 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 4 |
Hb_000645_120 |
0.0645011564 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 5 |
Hb_003531_060 |
0.0657797953 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000283_070 |
0.0711366482 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 7 |
Hb_009620_050 |
0.0718684132 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 8 |
Hb_000922_370 |
0.0728687845 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 9 |
Hb_000071_030 |
0.0733307188 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 10 |
Hb_001454_280 |
0.0735482711 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 11 |
Hb_011030_010 |
0.0789423749 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000107_500 |
0.0792426791 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 9 [Jatropha curcas] |
| 13 |
Hb_000983_150 |
0.0794758614 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 14 |
Hb_002235_160 |
0.0810179056 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 15 |
Hb_002174_020 |
0.0810330957 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_010193_050 |
0.0824188286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000088_130 |
0.0825786903 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 18 |
Hb_033187_050 |
0.0826580581 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 19 |
Hb_001140_350 |
0.0828947198 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 20 |
Hb_000680_090 |
0.0833628731 |
- |
- |
Protein SEY1, putative [Ricinus communis] |