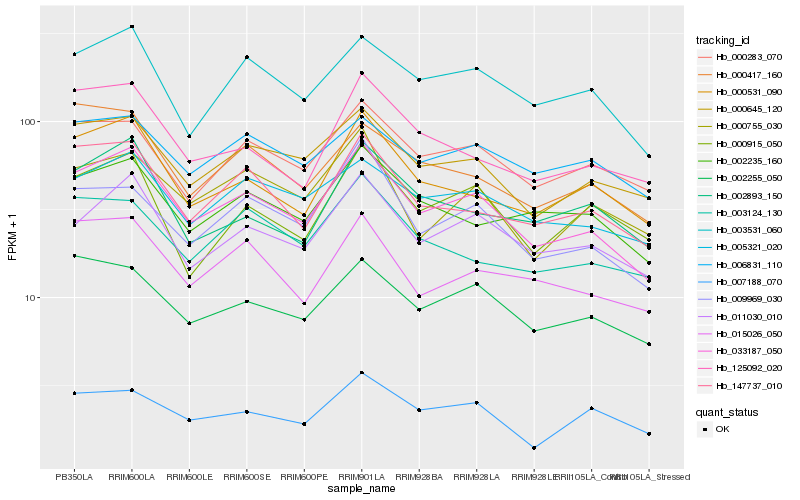
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033187_050 |
0.0 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 2 |
Hb_000645_120 |
0.0727991067 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 3 |
Hb_011030_010 |
0.0731597373 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_147737_010 |
0.0768761094 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003531_060 |
0.0773406533 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000531_090 |
0.0795751294 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 7 |
Hb_009969_030 |
0.0812000407 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005321_020 |
0.0826580581 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 9 |
Hb_002255_050 |
0.083496922 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_015026_050 |
0.0855491377 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_002235_160 |
0.0859722853 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 12 |
Hb_000283_070 |
0.0875092101 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 13 |
Hb_000755_030 |
0.0895598731 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 14 |
Hb_125092_020 |
0.0916482135 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_006831_110 |
0.0927367639 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 16 |
Hb_003124_130 |
0.0939487762 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000417_160 |
0.0960591545 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 18 |
Hb_007188_070 |
0.0961606449 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 19 |
Hb_002893_150 |
0.0970327051 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 20 |
Hb_000915_050 |
0.0981852467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |