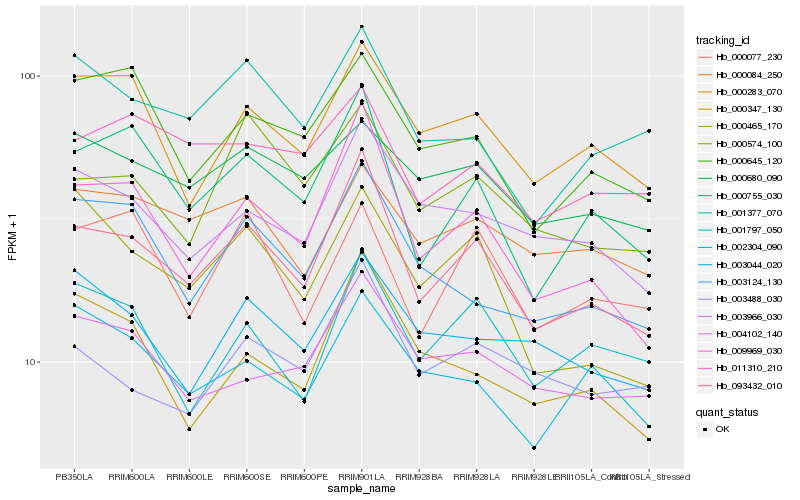
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093432_010 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 2 |
Hb_009969_030 |
0.0542428773 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000755_030 |
0.0702771812 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 4 |
Hb_003966_030 |
0.0874357228 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 5 |
Hb_000283_070 |
0.0877173622 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 6 |
Hb_000574_100 |
0.0899861419 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 7 |
Hb_003124_130 |
0.0936389797 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000645_120 |
0.0938316535 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 9 |
Hb_001377_070 |
0.0963198954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004102_140 |
0.0987651399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000084_250 |
0.0987824646 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 12 |
Hb_000347_130 |
0.098913491 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 13 |
Hb_011310_210 |
0.0997121975 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 14 |
Hb_000465_170 |
0.0999125664 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 15 |
Hb_003488_030 |
0.1001050564 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 16 |
Hb_001797_050 |
0.1005635784 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000680_090 |
0.1007590726 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 18 |
Hb_002304_090 |
0.1010319338 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003044_020 |
0.1012472473 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 20 |
Hb_000077_230 |
0.1018527222 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |