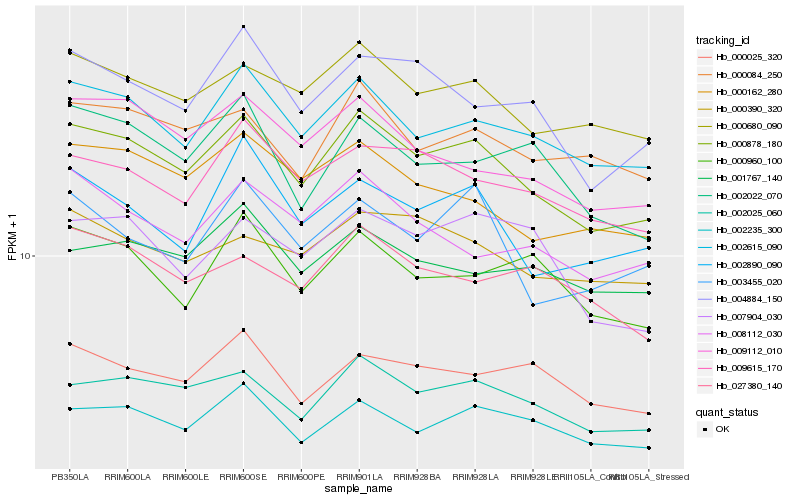
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_180 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002025_060 |
0.0518750221 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g73400, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002615_090 |
0.0562083325 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000680_090 |
0.058269738 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 5 |
Hb_002235_300 |
0.0686980985 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009615_170 |
0.0725290189 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000084_250 |
0.0726276842 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 8 |
Hb_000162_280 |
0.0726790895 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 9 |
Hb_009112_010 |
0.0727529111 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 10 |
Hb_004884_150 |
0.0743282804 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 11 |
Hb_000390_320 |
0.0792258613 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 12 |
Hb_002890_090 |
0.0802478864 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 13 |
Hb_000025_320 |
0.080373645 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 14 |
Hb_003455_020 |
0.0812073908 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 15 |
Hb_008112_030 |
0.0812434157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007904_030 |
0.0813789099 |
- |
- |
PREDICTED: nuclear mitotic apparatus protein 1 [Jatropha curcas] |
| 17 |
Hb_000960_100 |
0.0821464565 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 18 |
Hb_002022_070 |
0.0826098892 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_001767_140 |
0.0829620877 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 20 |
Hb_027380_140 |
0.083081894 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |