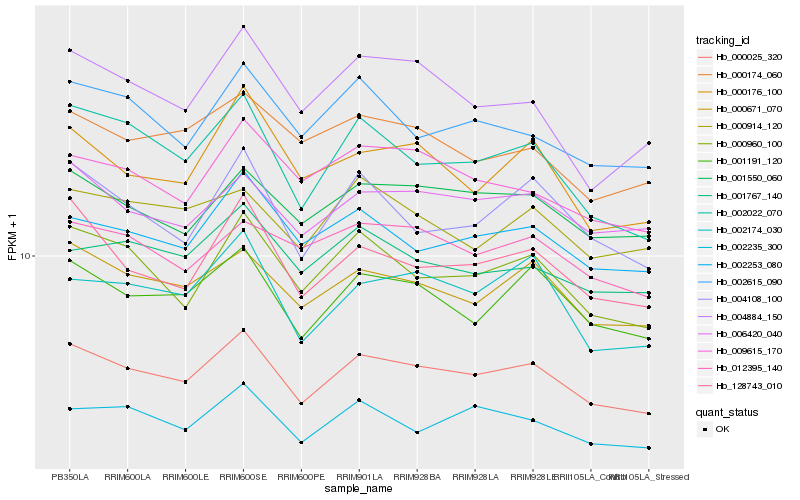
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_320 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 2 |
Hb_000671_070 |
0.0478595074 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 3 |
Hb_001191_120 |
0.0483540456 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001550_060 |
0.0548792053 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000960_100 |
0.05594227 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 6 |
Hb_002022_070 |
0.0561967029 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_006420_040 |
0.058404606 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_128743_010 |
0.0604536634 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 9 |
Hb_002253_080 |
0.0609478909 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 10 |
Hb_002235_300 |
0.0625449195 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004108_100 |
0.0630480304 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 12 |
Hb_002174_030 |
0.0641001699 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 13 |
Hb_009615_170 |
0.0652079708 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002615_090 |
0.0662642406 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012395_140 |
0.0675358665 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 16 |
Hb_000174_060 |
0.0678294909 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 17 |
Hb_001767_140 |
0.0687317904 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 18 |
Hb_000176_100 |
0.0692595156 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 19 |
Hb_004884_150 |
0.0695269586 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 20 |
Hb_000914_120 |
0.0702090306 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |