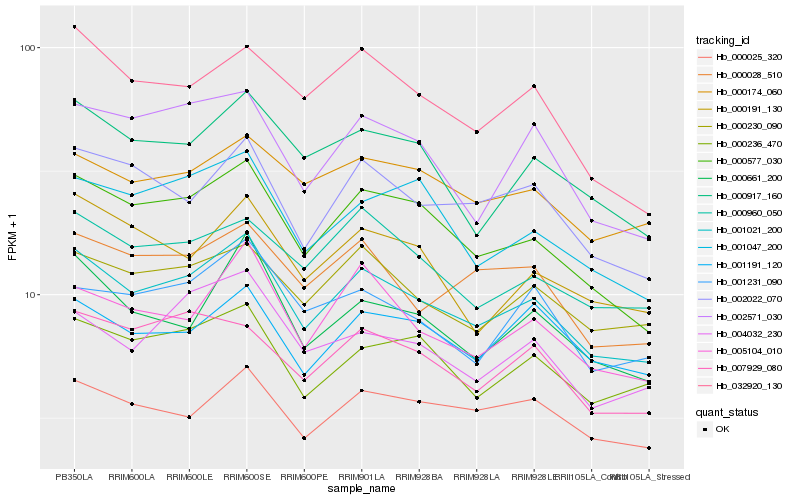
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_200 |
0.0 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 2 |
Hb_000577_030 |
0.0511501491 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 3 |
Hb_000230_090 |
0.0637460722 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002022_070 |
0.0658993336 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 5 |
Hb_005104_010 |
0.0676774671 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 6 |
Hb_000917_160 |
0.0681689361 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000236_470 |
0.0685976953 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 8 |
Hb_000661_200 |
0.0698285762 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000960_050 |
0.070124231 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 10 |
Hb_000025_320 |
0.0717255584 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 11 |
Hb_000028_510 |
0.0721637769 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 12 |
Hb_000174_060 |
0.072656032 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 13 |
Hb_004032_230 |
0.0728826117 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 14 |
Hb_032920_130 |
0.0731519235 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 15 |
Hb_002571_030 |
0.0733033774 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 16 |
Hb_001047_200 |
0.0737357682 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 17 |
Hb_007929_080 |
0.0750702006 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 18 |
Hb_001231_090 |
0.0754685696 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001191_120 |
0.0770608641 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000191_130 |
0.0779638503 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |