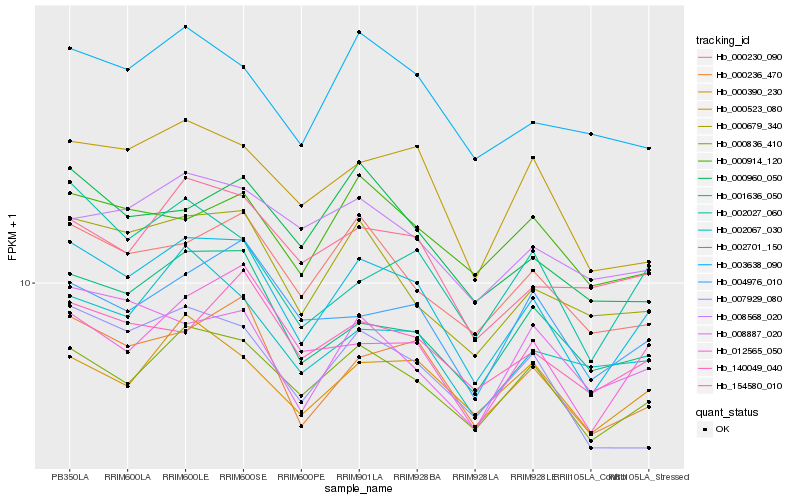
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002701_150 |
0.0 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000236_470 |
0.0576448919 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 3 |
Hb_000836_410 |
0.0592005671 |
- |
- |
sec10, putative [Ricinus communis] |
| 4 |
Hb_001636_050 |
0.0681410753 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 5 |
Hb_000679_340 |
0.0700298638 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000230_090 |
0.0706363239 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012565_050 |
0.0716733894 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 8 |
Hb_000523_080 |
0.0726663463 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_007929_080 |
0.0729081046 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 10 |
Hb_002027_060 |
0.0746384559 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 11 |
Hb_154580_010 |
0.075815579 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 12 |
Hb_004976_010 |
0.0765859771 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 13 |
Hb_003638_090 |
0.076880184 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 14 |
Hb_000390_230 |
0.0776739325 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 15 |
Hb_002067_030 |
0.0786047662 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_140049_040 |
0.078817428 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 17 |
Hb_008887_020 |
0.0803064662 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 18 |
Hb_000960_050 |
0.0822678571 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 19 |
Hb_000914_120 |
0.0826670047 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008568_020 |
0.0828812328 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |