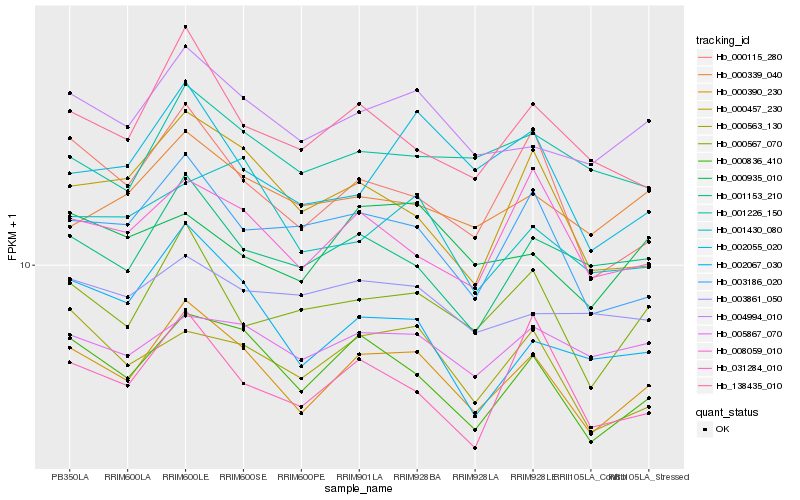
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_230 |
0.0 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 2 |
Hb_000836_410 |
0.0473026771 |
- |
- |
sec10, putative [Ricinus communis] |
| 3 |
Hb_004994_010 |
0.051804851 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_000115_280 |
0.0607048345 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 5 |
Hb_000567_070 |
0.0612218945 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 6 |
Hb_000563_130 |
0.0664522931 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_138435_010 |
0.0667705446 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 8 |
Hb_008059_010 |
0.0671706035 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001153_210 |
0.0671942734 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003186_020 |
0.0684881522 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005867_070 |
0.0713874934 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_031284_010 |
0.0723882032 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 13 |
Hb_001226_150 |
0.0727699221 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_002067_030 |
0.0731373168 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000935_010 |
0.073249633 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 16 |
Hb_000339_040 |
0.0740308016 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_001430_080 |
0.0743913409 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 18 |
Hb_002055_020 |
0.0751601533 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 19 |
Hb_000457_230 |
0.0756961302 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003861_050 |
0.0763147092 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |