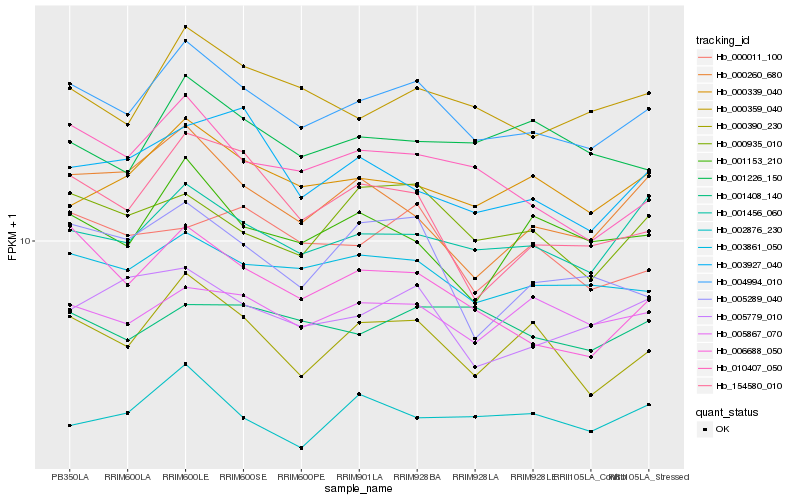
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_010 |
0.0 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_003861_050 |
0.0475862382 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 3 |
Hb_000390_230 |
0.051804851 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 4 |
Hb_001456_060 |
0.0537956222 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 5 |
Hb_000359_040 |
0.0543695791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010407_050 |
0.0578534011 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 7 |
Hb_000339_040 |
0.0602894339 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_005867_070 |
0.0607600067 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_006688_050 |
0.0622354208 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000935_010 |
0.0625105995 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 11 |
Hb_005779_010 |
0.0626880836 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_154580_010 |
0.0671992918 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 13 |
Hb_001153_210 |
0.0672359064 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000260_680 |
0.0686044546 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 15 |
Hb_001226_150 |
0.0687850549 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_002876_230 |
0.0688094862 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 17 |
Hb_000011_100 |
0.0694031589 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 18 |
Hb_001408_140 |
0.0702189695 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 19 |
Hb_005289_040 |
0.0702313448 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 20 |
Hb_003927_040 |
0.0708086946 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |