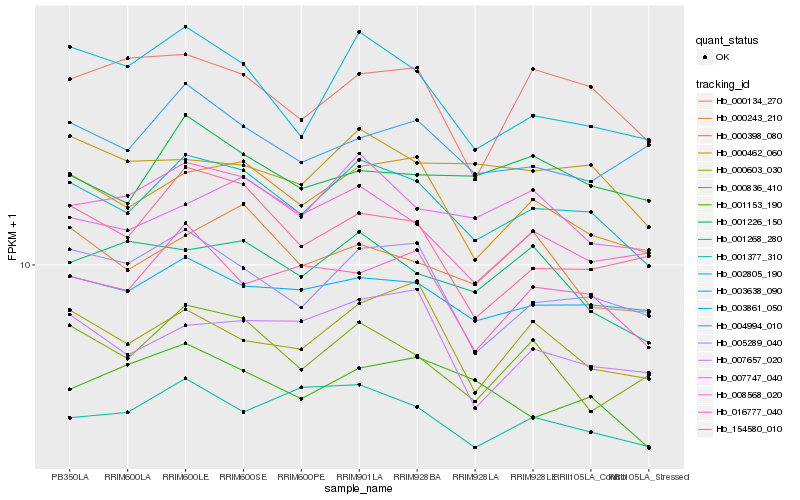
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002805_190 |
0.0 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 2 |
Hb_000398_080 |
0.0469328932 |
- |
- |
tip120, putative [Ricinus communis] |
| 3 |
Hb_003861_050 |
0.0590450373 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 4 |
Hb_005289_040 |
0.0622098833 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 5 |
Hb_003638_090 |
0.064669314 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 6 |
Hb_008568_020 |
0.0665608853 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 7 |
Hb_001377_310 |
0.0666971424 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 8 |
Hb_016777_040 |
0.067961314 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 9 |
Hb_000462_060 |
0.0708647843 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000836_410 |
0.0726061966 |
- |
- |
sec10, putative [Ricinus communis] |
| 11 |
Hb_001268_280 |
0.0735673675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000603_030 |
0.0747133886 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 13 |
Hb_154580_010 |
0.0748092625 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 14 |
Hb_007747_040 |
0.0749992199 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 15 |
Hb_000243_210 |
0.0751485927 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000134_270 |
0.0753610442 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 17 |
Hb_001153_190 |
0.0755149307 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 18 |
Hb_004994_010 |
0.0759146124 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_001226_150 |
0.0760167754 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_007657_020 |
0.0761266583 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |