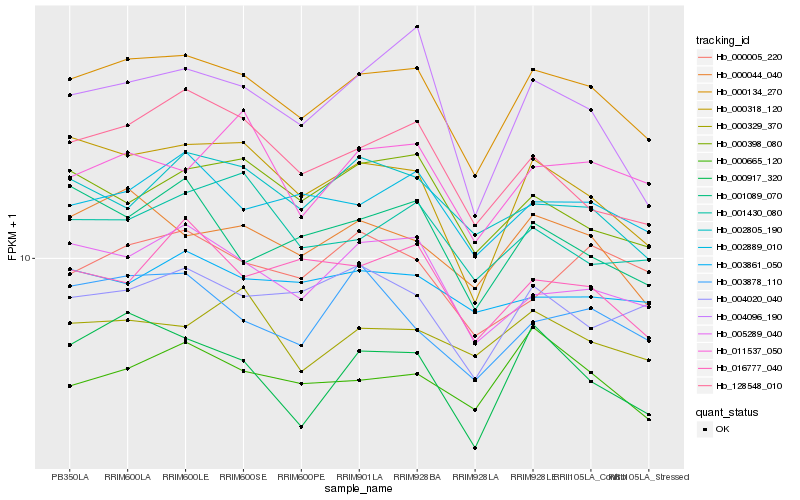
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_270 |
0.0 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 2 |
Hb_000044_040 |
0.0599714539 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 3 |
Hb_000318_120 |
0.0602776925 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005289_040 |
0.0640622275 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 5 |
Hb_002889_010 |
0.0662091905 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 6 |
Hb_000665_120 |
0.0728852458 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 7 |
Hb_004020_040 |
0.0744098228 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000917_320 |
0.0753412859 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002805_190 |
0.0753610442 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 10 |
Hb_000398_080 |
0.0759846409 |
- |
- |
tip120, putative [Ricinus communis] |
| 11 |
Hb_128548_010 |
0.0766970668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000329_370 |
0.0775704406 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 13 |
Hb_003861_050 |
0.0775863615 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 14 |
Hb_003878_110 |
0.0777256593 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 15 |
Hb_001089_070 |
0.078683283 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_011537_050 |
0.078954472 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001430_080 |
0.0790042915 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 18 |
Hb_016777_040 |
0.0791179735 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 19 |
Hb_004096_190 |
0.0793178739 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 20 |
Hb_000005_220 |
0.0795476517 |
- |
- |
DNA ligase 1, partial [Glycine soja] |