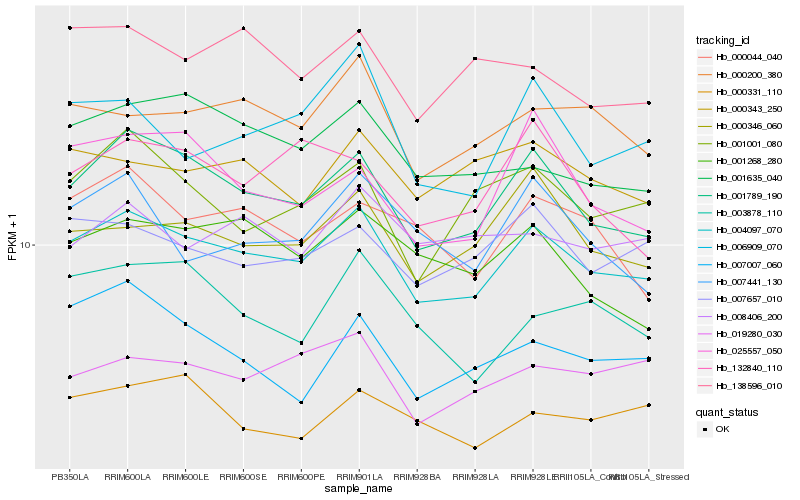
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004097_070 |
0.0 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 2 |
Hb_001789_190 |
0.0380910407 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 3 |
Hb_000200_380 |
0.0607215345 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007657_010 |
0.0636137616 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000346_060 |
0.0638410221 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 6 |
Hb_019280_030 |
0.0659678737 |
- |
- |
- |
| 7 |
Hb_001635_040 |
0.0680833435 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_001001_080 |
0.0686095052 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 9 |
Hb_000343_250 |
0.070403792 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 10 |
Hb_006909_070 |
0.0708754706 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_138596_010 |
0.0724340298 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 12 |
Hb_000044_040 |
0.0726770364 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 13 |
Hb_001268_280 |
0.0729041668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000331_110 |
0.073983833 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 15 |
Hb_025557_050 |
0.074581445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 16 |
Hb_008406_200 |
0.074959357 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007441_130 |
0.0754802763 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_007007_060 |
0.0755127731 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003878_110 |
0.0755171954 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 20 |
Hb_132840_110 |
0.0770646988 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |