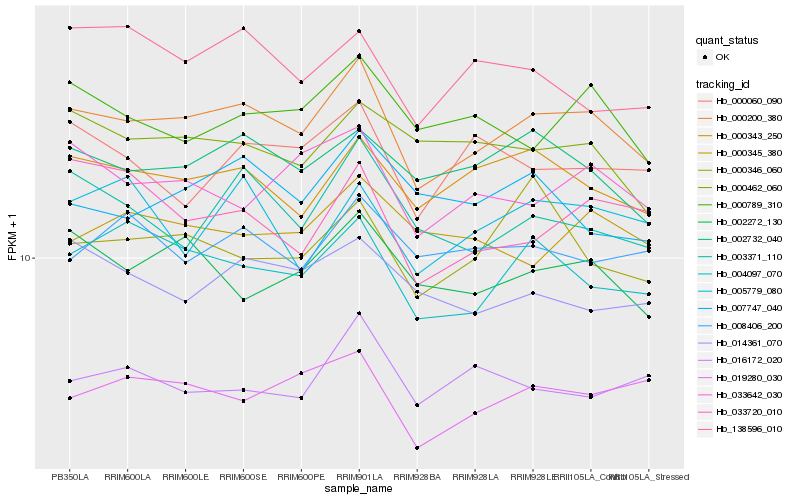
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000343_250 |
0.0593929923 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 3 |
Hb_004097_070 |
0.0607215345 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 4 |
Hb_002732_040 |
0.0622878926 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_019280_030 |
0.0664206702 |
- |
- |
- |
| 6 |
Hb_033642_030 |
0.0668964728 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002272_130 |
0.0682498788 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 8 |
Hb_003371_110 |
0.0683968667 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 9 |
Hb_000789_310 |
0.0705388389 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT1 [Jatropha curcas] |
| 10 |
Hb_000462_060 |
0.0733256146 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 11 |
Hb_014361_070 |
0.0739122192 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 12 |
Hb_008406_200 |
0.0746066123 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000060_090 |
0.0747682244 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a [Jatropha curcas] |
| 14 |
Hb_138596_010 |
0.0751835825 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 15 |
Hb_000346_060 |
0.0756260066 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 16 |
Hb_000345_380 |
0.0758867927 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 17 |
Hb_007747_040 |
0.076713575 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 18 |
Hb_016172_020 |
0.0771216417 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_005779_080 |
0.0775846556 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_033720_010 |
0.0779060471 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |