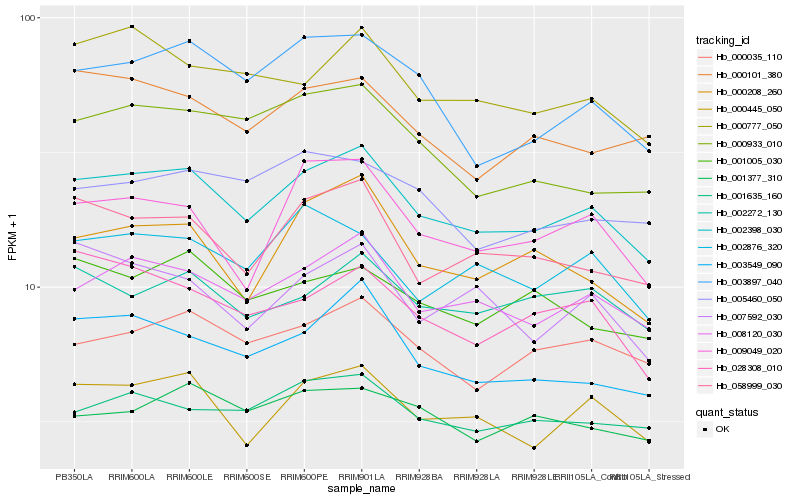
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002398_030 |
0.0 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_008120_030 |
0.0452110531 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 3 |
Hb_000445_050 |
0.0540602056 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 4 |
Hb_003549_090 |
0.0607202687 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 5 |
Hb_058999_030 |
0.0608073037 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_000035_110 |
0.0647491226 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 7 |
Hb_001635_160 |
0.0681680191 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 8 |
Hb_003897_040 |
0.0682995192 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 9 |
Hb_002272_130 |
0.0684736808 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 10 |
Hb_000777_050 |
0.0686584236 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 11 |
Hb_000208_260 |
0.0693554084 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 12 |
Hb_009049_020 |
0.0697458215 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 13 |
Hb_002876_320 |
0.0704852725 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 14 |
Hb_028308_010 |
0.0722376806 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001005_030 |
0.0728749938 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000101_380 |
0.0733217158 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 17 |
Hb_007592_030 |
0.073626751 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_000933_010 |
0.0739222846 |
- |
- |
- |
| 19 |
Hb_001377_310 |
0.0748644345 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 20 |
Hb_005460_050 |
0.0755313598 |
- |
- |
COR413-PM2, putative [Ricinus communis] |