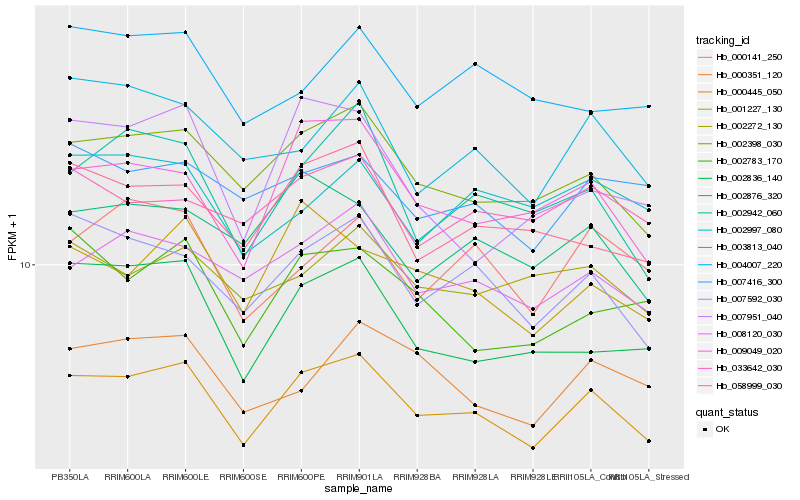
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_050 |
0.0 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_002398_030 |
0.0540602056 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_007592_030 |
0.0655761564 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 4 |
Hb_008120_030 |
0.0703834558 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 5 |
Hb_009049_020 |
0.0729411802 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 6 |
Hb_002942_060 |
0.0781582805 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 7 |
Hb_058999_030 |
0.078905603 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_001227_130 |
0.0801747784 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 9 |
Hb_002836_140 |
0.0802115067 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 10 |
Hb_002876_320 |
0.0809860032 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 11 |
Hb_003813_040 |
0.0818917847 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 12 |
Hb_000351_120 |
0.0849455219 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 13 |
Hb_007416_300 |
0.0849780602 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 14 |
Hb_000141_250 |
0.0852622331 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X3 [Jatropha curcas] |
| 15 |
Hb_002783_170 |
0.0857987054 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 16 |
Hb_002272_130 |
0.0865544877 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 17 |
Hb_033642_030 |
0.0898228686 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004007_220 |
0.0917464715 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 19 |
Hb_007951_040 |
0.0918271691 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 20 |
Hb_002997_080 |
0.0936905264 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |