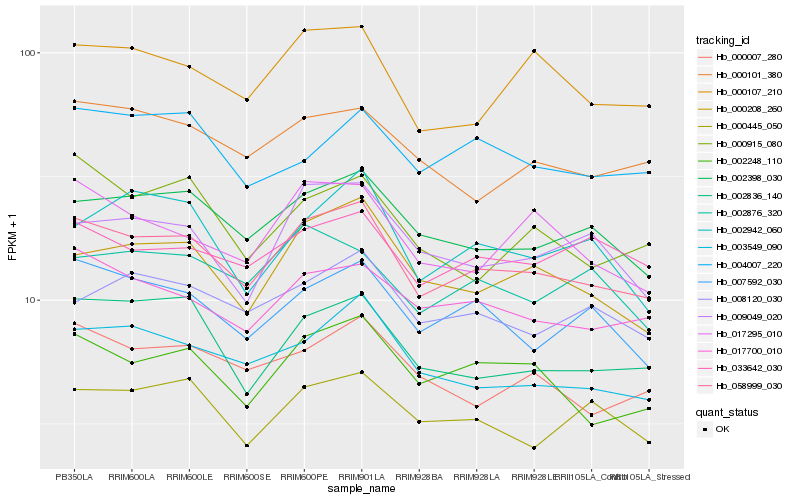
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058999_030 |
0.0 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_002398_030 |
0.0608073037 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002248_110 |
0.0629521013 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 4 |
Hb_017700_010 |
0.0650124277 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 5 |
Hb_003549_090 |
0.066961321 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 6 |
Hb_000208_260 |
0.0676467403 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_007592_030 |
0.0701087174 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_000107_210 |
0.0707298502 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 9 |
Hb_002836_140 |
0.0739081726 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 10 |
Hb_000007_280 |
0.0749015145 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 11 |
Hb_000101_380 |
0.0755191593 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 12 |
Hb_017295_010 |
0.0758736797 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 13 |
Hb_008120_030 |
0.0767367366 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 14 |
Hb_009049_020 |
0.0784811109 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 15 |
Hb_000445_050 |
0.078905603 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_033642_030 |
0.0792178063 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002876_320 |
0.080999846 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 18 |
Hb_004007_220 |
0.0836418773 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 19 |
Hb_000915_080 |
0.0842053506 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 20 |
Hb_002942_060 |
0.0849033284 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |