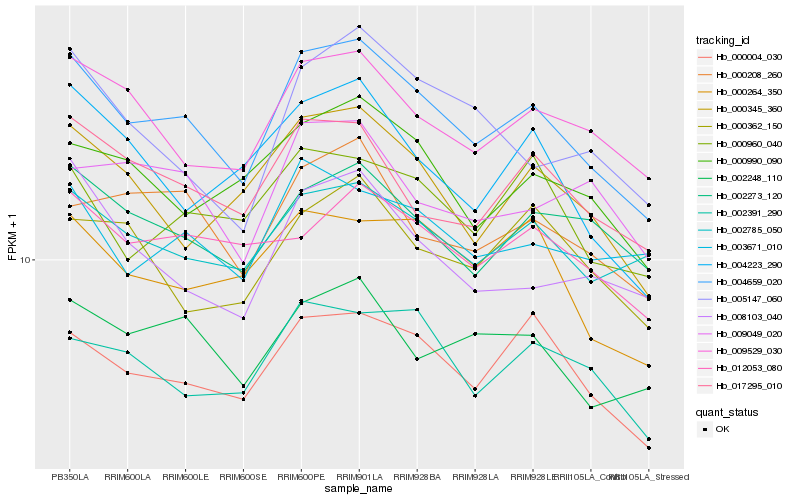
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004659_020 |
0.0 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 2 |
Hb_000004_030 |
0.075814534 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 3 |
Hb_009529_030 |
0.0801369659 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 4 |
Hb_017295_010 |
0.0840692577 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 5 |
Hb_002785_050 |
0.0860025307 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 6 |
Hb_002248_110 |
0.0866622497 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 7 |
Hb_002273_120 |
0.0872442996 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 8 |
Hb_000345_360 |
0.0877950854 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 9 |
Hb_000208_260 |
0.0896797273 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000990_090 |
0.0898969324 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_002391_290 |
0.0917169685 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000264_350 |
0.0924437934 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 13 |
Hb_003671_010 |
0.0930038145 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000960_040 |
0.0930470152 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 15 |
Hb_008103_040 |
0.0931181839 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_004223_290 |
0.0941592254 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 17 |
Hb_012053_080 |
0.0953322403 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 18 |
Hb_005147_060 |
0.0981400792 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 19 |
Hb_009049_020 |
0.1012711003 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 20 |
Hb_000362_150 |
0.1013552787 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |