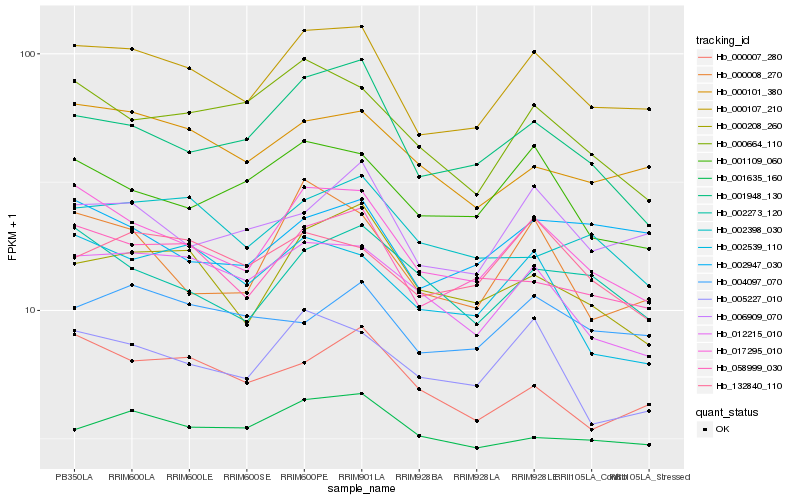
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_210 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_017295_010 |
0.0578456153 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 3 |
Hb_058999_030 |
0.0707298502 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_006909_070 |
0.0740712249 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_000101_380 |
0.0780731541 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_005227_010 |
0.0810098625 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012215_010 |
0.08193141 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 8 |
Hb_001109_060 |
0.0837901616 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_004097_070 |
0.0839552995 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 10 |
Hb_001948_130 |
0.0852733711 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 11 |
Hb_132840_110 |
0.0853636713 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_001635_160 |
0.0866658222 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 13 |
Hb_000208_260 |
0.0873321155 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_000007_280 |
0.08776155 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 15 |
Hb_000008_270 |
0.0877744486 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein REVOLUTA [Jatropha curcas] |
| 16 |
Hb_002947_030 |
0.0881748272 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 17 |
Hb_000664_110 |
0.0897579482 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 18 |
Hb_002273_120 |
0.0927030191 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 19 |
Hb_002398_030 |
0.0943683094 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002539_110 |
0.0945629234 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |