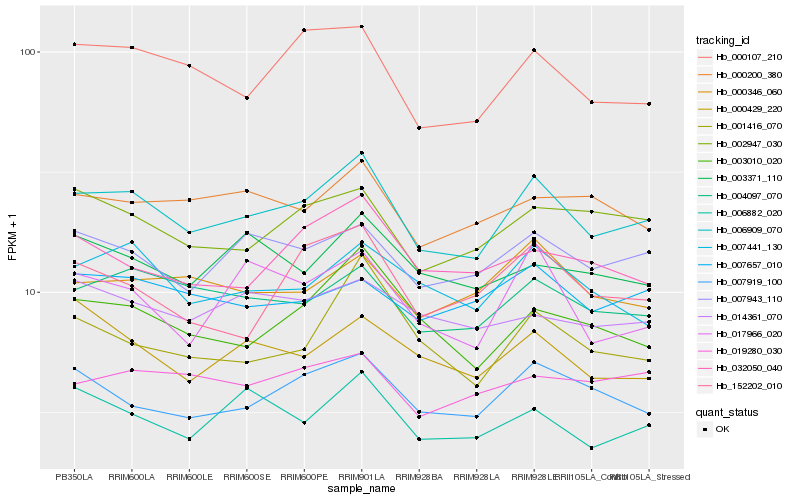
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006909_070 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_004097_070 |
0.0708754706 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 3 |
Hb_000107_210 |
0.0740712249 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_007943_110 |
0.0743421371 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 5 |
Hb_007919_100 |
0.0777332241 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 6 |
Hb_002947_030 |
0.0807140785 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 7 |
Hb_007441_130 |
0.0815687204 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003010_020 |
0.0815971654 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000346_060 |
0.0850307838 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 10 |
Hb_006882_020 |
0.0881015259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000200_380 |
0.0881714693 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007657_010 |
0.0888917387 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_032050_040 |
0.0891771201 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 14 |
Hb_019280_030 |
0.0895892156 |
- |
- |
- |
| 15 |
Hb_017966_020 |
0.0901052219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03560, mitochondrial [Jatropha curcas] |
| 16 |
Hb_152202_010 |
0.0913324293 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 17 |
Hb_001416_070 |
0.0914965719 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 18 |
Hb_003371_110 |
0.0919414639 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 19 |
Hb_000429_220 |
0.0931910903 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 20 |
Hb_014361_070 |
0.0934995234 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |