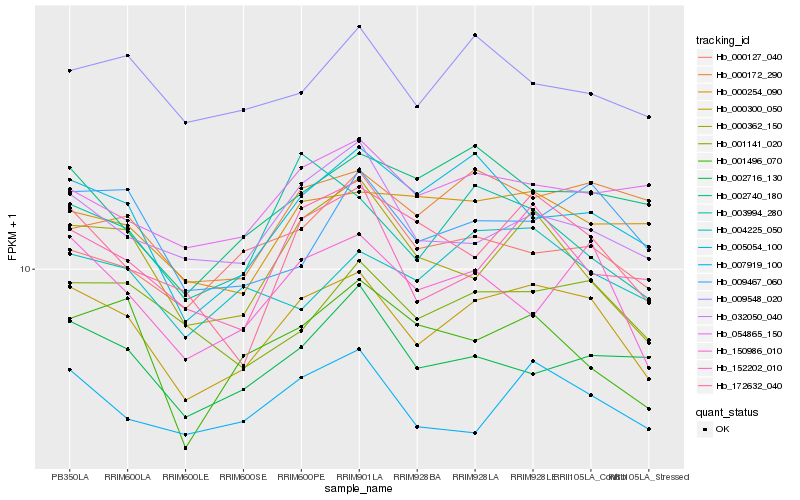
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_150986_010 |
0.0836290781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007919_100 |
0.0862183124 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 4 |
Hb_000362_150 |
0.0864335645 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 5 |
Hb_005054_100 |
0.086551982 |
- |
- |
PREDICTED: uncharacterized protein At2g39920 [Jatropha curcas] |
| 6 |
Hb_001141_020 |
0.0887550384 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 7 |
Hb_152202_010 |
0.0897490542 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 8 |
Hb_003994_280 |
0.0915289804 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 9 |
Hb_032050_040 |
0.0917535578 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 10 |
Hb_009467_060 |
0.0933944704 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_172632_040 |
0.0935788048 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 12 |
Hb_000254_090 |
0.0939674219 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 13 |
Hb_004225_050 |
0.0947128444 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_009548_020 |
0.0953858943 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 15 |
Hb_002740_180 |
0.0962846797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000127_040 |
0.0971399803 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 17 |
Hb_054865_150 |
0.0972815369 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein Q [Jatropha curcas] |
| 18 |
Hb_002716_130 |
0.0984637742 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001496_070 |
0.0990591899 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_000172_290 |
0.0997450833 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |