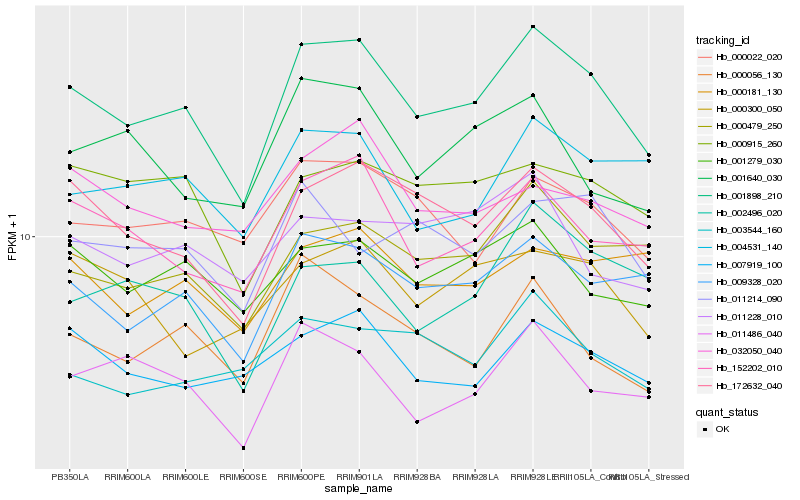
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_210 |
0.0 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 2 |
Hb_172632_040 |
0.0759209527 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 3 |
Hb_009328_020 |
0.0816355558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_152202_010 |
0.0852260773 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 5 |
Hb_000479_250 |
0.0884253846 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 6 |
Hb_001279_030 |
0.0934345096 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_000022_020 |
0.0965930784 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 8 |
Hb_000056_130 |
0.0971129141 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 9 |
Hb_011486_040 |
0.0975702662 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 10 |
Hb_007919_100 |
0.0984309126 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 11 |
Hb_004531_140 |
0.0985412106 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 12 |
Hb_000915_260 |
0.0996879681 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002496_020 |
0.1041445671 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000181_130 |
0.1051115543 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_001640_030 |
0.1066413734 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 16 |
Hb_003544_160 |
0.1077214584 |
- |
- |
- |
| 17 |
Hb_000300_050 |
0.1078465761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011214_090 |
0.1082563899 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011228_010 |
0.1091411721 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_032050_040 |
0.1092411906 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |