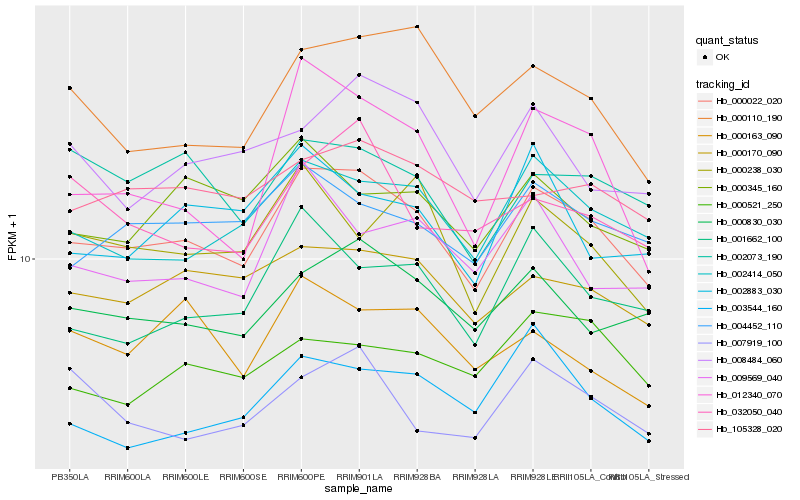
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 2 |
Hb_002414_050 |
0.0628565299 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 3 |
Hb_000830_030 |
0.0691016513 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 4 |
Hb_000170_090 |
0.070203627 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 5 |
Hb_004452_110 |
0.0732183617 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001662_100 |
0.075993882 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 7 |
Hb_000345_160 |
0.0811559525 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 8 |
Hb_105328_020 |
0.0817342051 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 9 |
Hb_000110_190 |
0.082374148 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 10 |
Hb_002073_190 |
0.0833979588 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009569_040 |
0.0835987908 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 12 |
Hb_008484_060 |
0.0846989524 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 13 |
Hb_003544_160 |
0.0850542411 |
- |
- |
- |
| 14 |
Hb_012340_070 |
0.0852568837 |
- |
- |
Gamma-secretase subunit APH1-like family protein [Populus trichocarpa] |
| 15 |
Hb_000238_030 |
0.0854935836 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_002883_030 |
0.0866287134 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 17 |
Hb_032050_040 |
0.0869416509 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 18 |
Hb_007919_100 |
0.0870149853 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 19 |
Hb_000521_250 |
0.0877027896 |
- |
- |
PREDICTED: protein N-terminal asparagine amidohydrolase isoform X2 [Jatropha curcas] |
| 20 |
Hb_000163_090 |
0.0878132266 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |