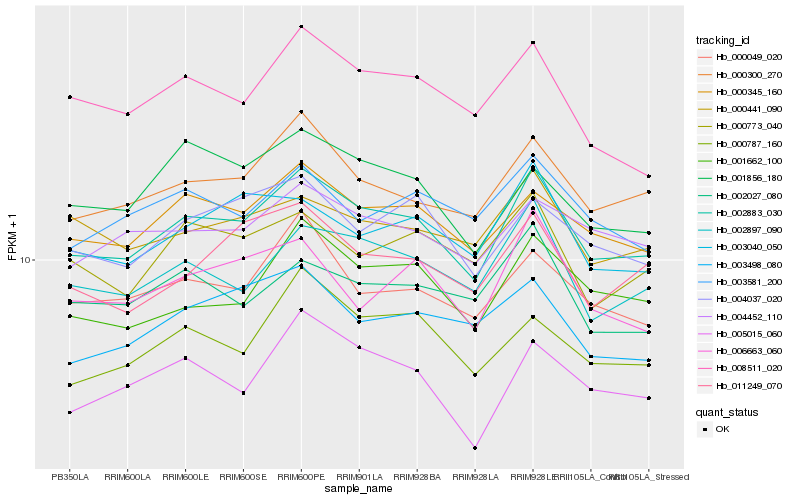
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002883_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 2 |
Hb_000345_160 |
0.0557854273 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 3 |
Hb_000300_270 |
0.0559592359 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_002897_090 |
0.0592105994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001662_100 |
0.0627626059 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 6 |
Hb_000049_020 |
0.0658001126 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 7 |
Hb_004037_020 |
0.0692324697 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008511_020 |
0.0693067382 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 9 |
Hb_004452_110 |
0.0699829758 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003040_050 |
0.0703148937 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 11 |
Hb_011249_070 |
0.0708784267 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 12 |
Hb_000787_160 |
0.0715590989 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000773_040 |
0.0726630949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005015_060 |
0.0732821203 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 15 |
Hb_002027_080 |
0.0735510925 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 16 |
Hb_000441_090 |
0.073777282 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 17 |
Hb_006663_060 |
0.0743927563 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 18 |
Hb_003498_080 |
0.0748290548 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_001856_180 |
0.0751874847 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 20 |
Hb_003581_200 |
0.0753833226 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |