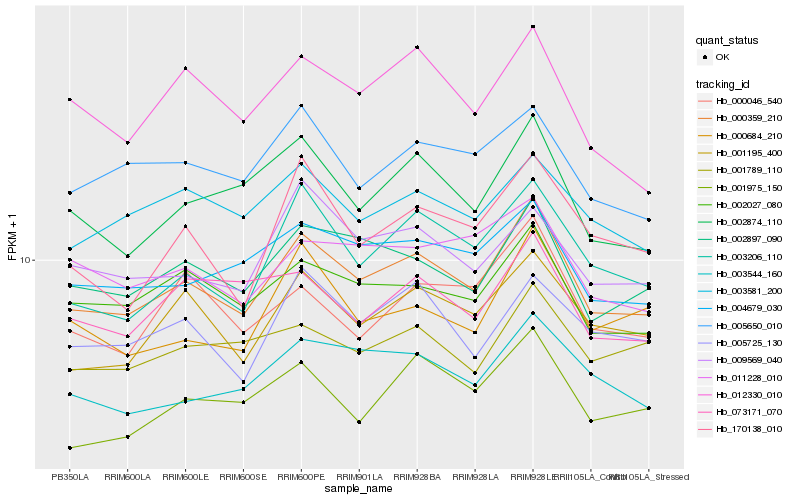
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002897_090 |
0.0606076846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002027_080 |
0.0630311484 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 4 |
Hb_001195_400 |
0.0656201612 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 5 |
Hb_002874_110 |
0.066862158 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 6 |
Hb_004679_030 |
0.0669182444 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 7 |
Hb_003206_110 |
0.0695128159 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 8 |
Hb_003544_160 |
0.0705103177 |
- |
- |
- |
| 9 |
Hb_005650_010 |
0.0745254397 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_003581_200 |
0.0758597511 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 11 |
Hb_000684_210 |
0.0764505549 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 12 |
Hb_005725_130 |
0.0773347753 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_009569_040 |
0.0778124548 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 14 |
Hb_170138_010 |
0.0782359998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012330_010 |
0.0783724465 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001975_150 |
0.078708096 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 17 |
Hb_001789_110 |
0.0791468353 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_011228_010 |
0.0799188882 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000046_540 |
0.0807086314 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 20 |
Hb_073171_070 |
0.0814089252 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |