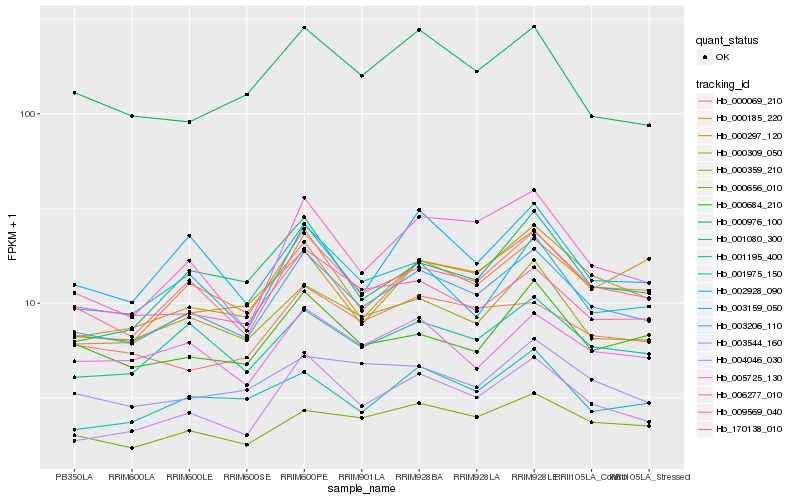
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003206_110 |
0.0 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 2 |
Hb_170138_010 |
0.045883645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004046_030 |
0.0517741641 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 4 |
Hb_000185_220 |
0.0678151486 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 5 |
Hb_001195_400 |
0.0686097224 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 6 |
Hb_000359_210 |
0.0695128159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006277_010 |
0.0747693022 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 8 |
Hb_000656_010 |
0.0751972318 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000309_050 |
0.0800571524 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001975_150 |
0.0814852307 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 11 |
Hb_005725_130 |
0.0830557445 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_000976_100 |
0.0862919044 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 13 |
Hb_000684_210 |
0.0866627422 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 14 |
Hb_002928_090 |
0.0871094882 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 15 |
Hb_009569_040 |
0.0871623006 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 16 |
Hb_000069_210 |
0.0873311697 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 17 |
Hb_001080_300 |
0.0891326923 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 18 |
Hb_003544_160 |
0.0894437918 |
- |
- |
- |
| 19 |
Hb_000297_120 |
0.0898010842 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 20 |
Hb_003159_050 |
0.0910911166 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |