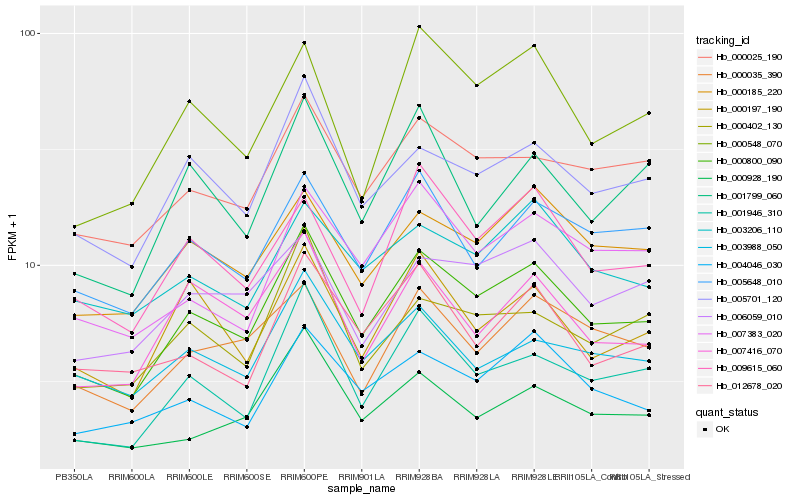
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000800_090 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 2 |
Hb_005701_120 |
0.0787946323 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 3 |
Hb_000025_190 |
0.0829618573 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_001946_310 |
0.0833299956 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 5 |
Hb_004046_030 |
0.0879134173 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 6 |
Hb_000185_220 |
0.0893869528 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 7 |
Hb_005648_010 |
0.0914015692 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 8 |
Hb_000197_190 |
0.0928703151 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000548_070 |
0.0933784532 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_007383_020 |
0.093934647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007416_070 |
0.0941711917 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 12 |
Hb_000928_190 |
0.0964510321 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000035_390 |
0.0972752855 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 14 |
Hb_012678_020 |
0.0973474828 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006059_010 |
0.0988996219 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 16 |
Hb_003206_110 |
0.0991246366 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 17 |
Hb_009615_060 |
0.0992532958 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 18 |
Hb_000402_130 |
0.0996252298 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 19 |
Hb_001799_060 |
0.0999897744 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 20 |
Hb_003988_050 |
0.1008094144 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |