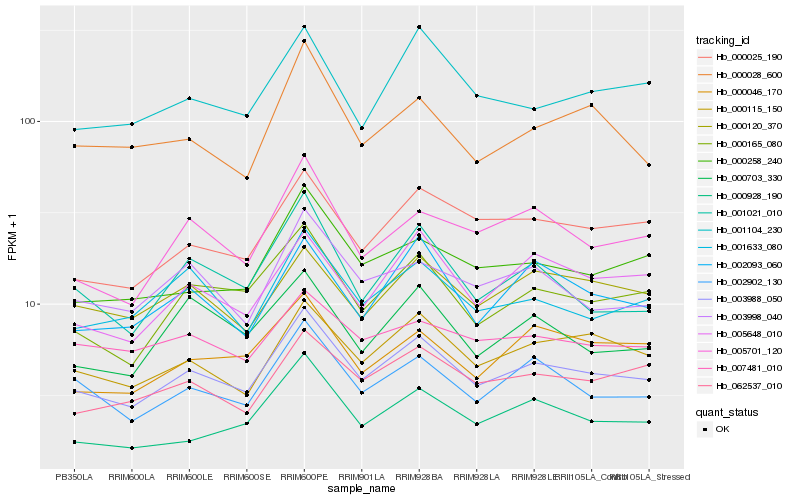
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003988_050 |
0.0 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 2 |
Hb_000115_150 |
0.0686177038 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 3 |
Hb_000165_080 |
0.0697174844 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002902_130 |
0.0720652274 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 5 |
Hb_001633_080 |
0.0779939107 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 6 |
Hb_000258_240 |
0.0782848373 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 7 |
Hb_000025_190 |
0.0784563162 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_005701_120 |
0.079080621 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 9 |
Hb_062537_010 |
0.0800020672 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 10 |
Hb_003998_040 |
0.0815737848 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 11 |
Hb_005648_010 |
0.0853014215 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 12 |
Hb_000120_370 |
0.0857886146 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 13 |
Hb_002093_060 |
0.0861015171 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_001021_010 |
0.0865801048 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 15 |
Hb_000046_170 |
0.0873449233 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007481_010 |
0.0900278396 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 17 |
Hb_000028_600 |
0.0902224374 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 18 |
Hb_000928_190 |
0.0905816768 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001104_230 |
0.0907829652 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 20 |
Hb_000703_330 |
0.091535943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |