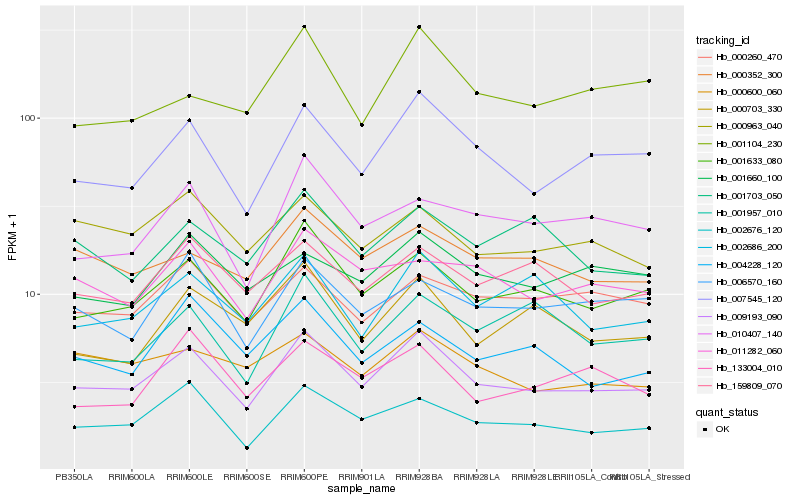
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_090 |
0.0 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007545_120 |
0.0701703946 |
- |
- |
NADH:cytochrome b5 reductase [Vernicia fordii] |
| 3 |
Hb_001633_080 |
0.0751893715 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 4 |
Hb_002676_120 |
0.0820772251 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 5 |
Hb_002686_200 |
0.1033284633 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 6 |
Hb_159809_070 |
0.1050662081 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 7 |
Hb_006570_160 |
0.1057597091 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 8 |
Hb_010407_140 |
0.1067943838 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 9 |
Hb_011282_060 |
0.107189765 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 10 |
Hb_000352_300 |
0.1086450701 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 11 |
Hb_000963_040 |
0.1089430598 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 12 |
Hb_000703_330 |
0.109825897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000260_470 |
0.1118064831 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 14 |
Hb_133004_010 |
0.1119007234 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 15 |
Hb_004228_120 |
0.1124058765 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 16 |
Hb_001660_100 |
0.1129544284 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 17 |
Hb_000600_060 |
0.1131193464 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001957_010 |
0.1134643472 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001104_230 |
0.1136877547 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 20 |
Hb_001703_050 |
0.1148892713 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |