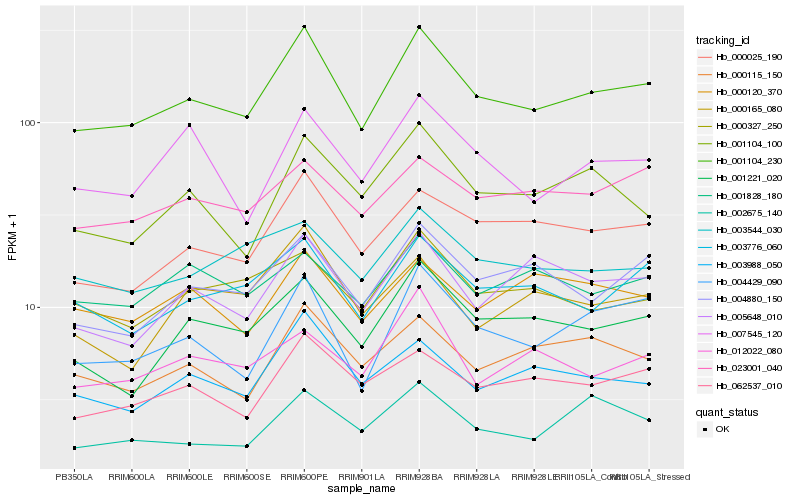
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_230 |
0.0 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 2 |
Hb_000025_190 |
0.0814955435 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_004429_090 |
0.0845053311 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001221_020 |
0.0859514755 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 5 |
Hb_003776_060 |
0.0872662915 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 6 |
Hb_062537_010 |
0.0887060778 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 7 |
Hb_004880_150 |
0.0892727529 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 8 |
Hb_003988_050 |
0.0907829652 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 9 |
Hb_000115_150 |
0.0951492762 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 10 |
Hb_000327_250 |
0.0998176049 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 11 |
Hb_003544_030 |
0.0998920688 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 12 |
Hb_001104_100 |
0.1004910593 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 13 |
Hb_023001_040 |
0.1007808181 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_005648_010 |
0.100969258 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 15 |
Hb_007545_120 |
0.1013941478 |
- |
- |
NADH:cytochrome b5 reductase [Vernicia fordii] |
| 16 |
Hb_002675_140 |
0.1048421841 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 17 |
Hb_000165_080 |
0.1087785419 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001828_180 |
0.1087914459 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 19 |
Hb_012022_080 |
0.1088523517 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000120_370 |
0.1094501556 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |