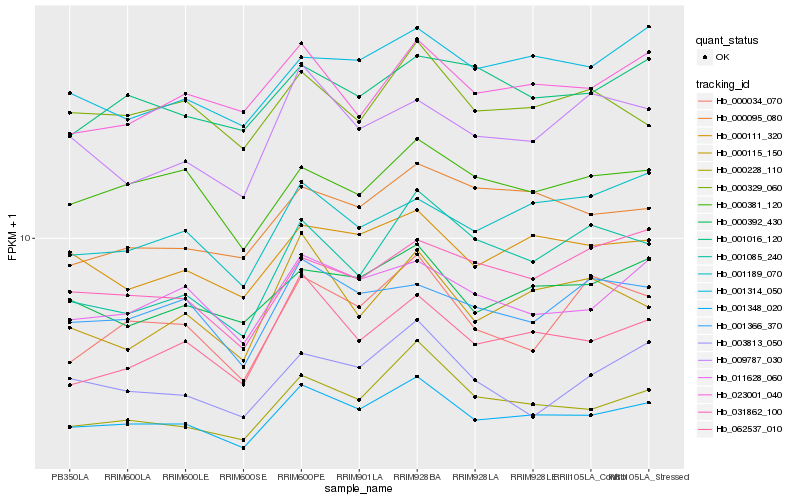
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 2 |
Hb_062537_010 |
0.0717292887 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 3 |
Hb_000228_110 |
0.0717352783 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_011628_060 |
0.0751143998 |
- |
- |
Adenosine 3'-phospho 5'-phosphosulfate transporter, putative [Ricinus communis] |
| 5 |
Hb_000111_320 |
0.0761916954 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 6 |
Hb_000392_430 |
0.076990016 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 7 |
Hb_023001_040 |
0.0875437912 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_001366_370 |
0.0878178654 |
- |
- |
PREDICTED: aarF domain-containing protein kinase 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001314_050 |
0.0881021067 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_000381_120 |
0.0885341602 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 11 |
Hb_000115_150 |
0.0886635934 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 12 |
Hb_000095_080 |
0.0901230279 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 13 |
Hb_000034_070 |
0.0902115855 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 14 |
Hb_000329_060 |
0.0903506028 |
- |
- |
chloroplast 5-enolpyruvylshikimate 3-phosphate synthase [Hevea brasiliensis] |
| 15 |
Hb_009787_030 |
0.0921715031 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 16 |
Hb_001085_240 |
0.093042045 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003813_050 |
0.0934057028 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001189_070 |
0.0943163787 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 19 |
Hb_031862_100 |
0.0946231051 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001016_120 |
0.0949160385 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |