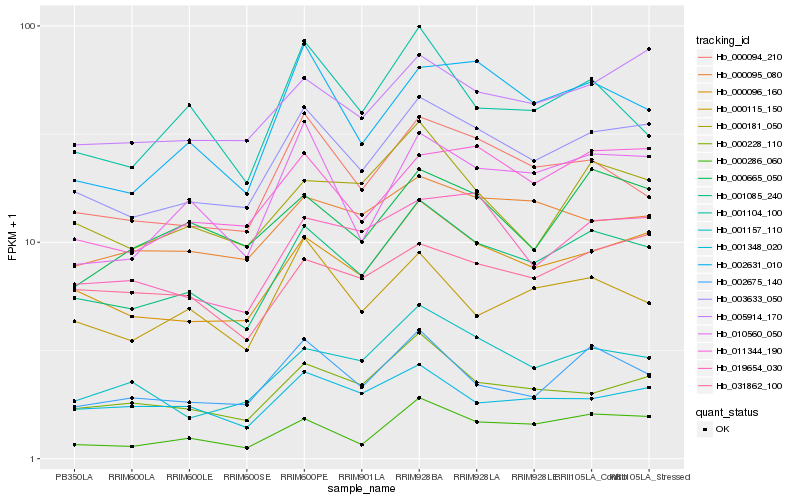
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_240 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003633_050 |
0.0492348557 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 3 |
Hb_000096_160 |
0.0605451192 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 4 |
Hb_000094_210 |
0.0849400317 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 5 |
Hb_000228_110 |
0.0859485859 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002675_140 |
0.0859709069 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 7 |
Hb_005914_170 |
0.091485926 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 8 |
Hb_001348_020 |
0.093042045 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 9 |
Hb_010560_050 |
0.0932190324 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 10 |
Hb_000095_080 |
0.0932895221 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |
| 11 |
Hb_000181_050 |
0.0939952687 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 12 |
Hb_011344_190 |
0.0940761754 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 13 |
Hb_019654_030 |
0.094120893 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_002631_010 |
0.0965145651 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 15 |
Hb_001157_110 |
0.0965198669 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 16 |
Hb_000115_150 |
0.0973137091 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 17 |
Hb_001104_100 |
0.0973868958 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 18 |
Hb_031862_100 |
0.0975988026 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000286_060 |
0.1000845231 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 20 |
Hb_000665_050 |
0.1002663322 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |