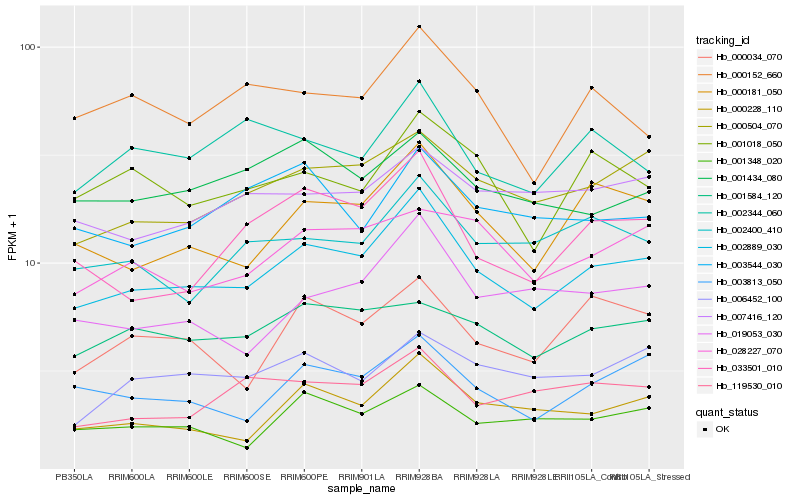
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002889_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 [Jatropha curcas] |
| 2 |
Hb_000181_050 |
0.0796477879 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 3 |
Hb_000504_070 |
0.0856873603 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000228_110 |
0.0895400336 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_003813_050 |
0.0927567161 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_033501_010 |
0.096106242 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 7 |
Hb_019053_030 |
0.0993376501 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 8 |
Hb_002400_410 |
0.1025978271 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 9 |
Hb_001018_050 |
0.1036195036 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_028227_070 |
0.104040694 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000034_070 |
0.1044972861 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 3B-like [Jatropha curcas] |
| 12 |
Hb_001348_020 |
0.1046366657 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001584_120 |
0.1072332884 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase 3 [Jatropha curcas] |
| 14 |
Hb_119530_010 |
0.1084705769 |
- |
- |
- |
| 15 |
Hb_006452_100 |
0.1087899986 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 16 |
Hb_007416_120 |
0.1088230357 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 17 |
Hb_000152_660 |
0.109105981 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 18 |
Hb_001434_080 |
0.1100656642 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 19 |
Hb_002344_060 |
0.1117639367 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 20 |
Hb_003544_030 |
0.1122288815 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |