| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
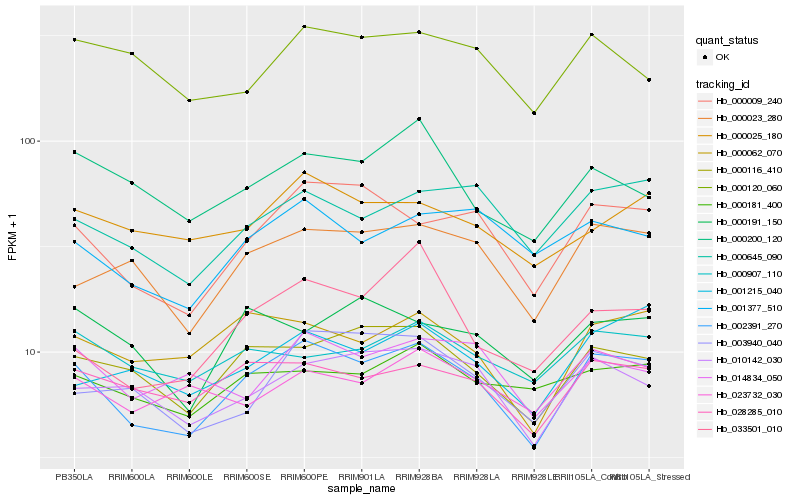
Hb_002391_270 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 2 |
Hb_000009_240 |
0.0842212237 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000116_410 |
0.0885914186 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 4 |
Hb_000645_090 |
0.095188974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_028285_010 |
0.0965120808 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 6 |
Hb_000023_280 |
0.0972656949 |
- |
- |
hypothetical protein POPTR_0006s28210g [Populus trichocarpa] |
| 7 |
Hb_010142_030 |
0.0992796513 |
- |
- |
PREDICTED: isoamylase 3, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000062_070 |
0.1006005411 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_001377_510 |
0.1040881888 |
- |
- |
ormdl, putative [Ricinus communis] |
| 10 |
Hb_003940_040 |
0.1069636927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000025_180 |
0.1088275489 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 12 |
Hb_000907_110 |
0.1109084163 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 13 |
Hb_001215_040 |
0.1110286442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000181_400 |
0.1117129319 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 15 |
Hb_014834_050 |
0.1122620495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000200_120 |
0.1130343511 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000191_150 |
0.1149540228 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 18 |
Hb_023732_030 |
0.1160044714 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000120_060 |
0.1170224273 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 20 |
Hb_033501_010 |
0.1173616933 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |