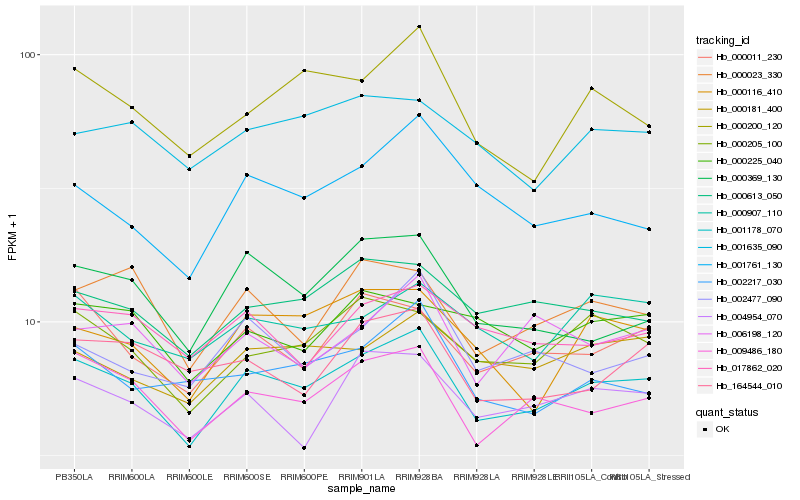
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001178_070 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 2 |
Hb_009486_180 |
0.0604636259 |
- |
- |
unknown [Glycine max] |
| 3 |
Hb_000205_100 |
0.0734937658 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 4 |
Hb_000011_230 |
0.0761750464 |
- |
- |
PREDICTED: bifunctional protein FolD 4, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000613_050 |
0.0765612874 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004954_070 |
0.0777828405 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 7 |
Hb_000181_400 |
0.0779895564 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000116_410 |
0.0785805555 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 9 |
Hb_000023_330 |
0.0794422137 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 10 |
Hb_000200_120 |
0.0818171315 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000907_110 |
0.0822538945 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 12 |
Hb_000369_130 |
0.0823986901 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 13 |
Hb_001635_090 |
0.0835884816 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 14 |
Hb_002477_090 |
0.0836833839 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 15 |
Hb_017862_020 |
0.0839162076 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 16 |
Hb_006198_120 |
0.0848333152 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 17 |
Hb_002217_030 |
0.0863444673 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_164544_010 |
0.0876605534 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000225_040 |
0.0877785377 |
- |
- |
PREDICTED: translation factor GUF1 homolog, mitochondrial [Nelumbo nucifera] |
| 20 |
Hb_001761_130 |
0.088267203 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |