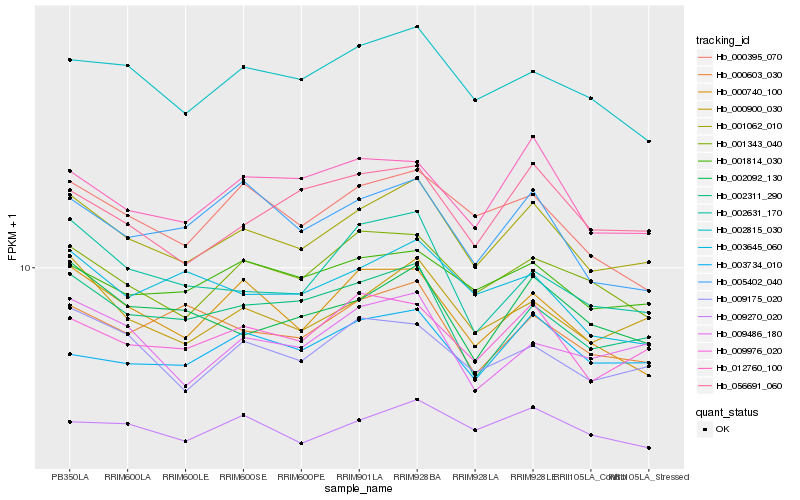
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001062_010 |
0.0 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 2 |
Hb_000900_030 |
0.0494022671 |
- |
- |
PREDICTED: WD repeat-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_009976_020 |
0.0584470478 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 4 |
Hb_001343_040 |
0.0623601552 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 5 |
Hb_002311_290 |
0.0636671781 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 6 |
Hb_002815_030 |
0.0639435064 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 7 |
Hb_009175_020 |
0.0646139139 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_001814_030 |
0.0661098122 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 9 |
Hb_002092_130 |
0.06613209 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 10 |
Hb_000740_100 |
0.0677438289 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_009270_020 |
0.0684582711 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 12 |
Hb_012760_100 |
0.0687079377 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 13 |
Hb_000395_070 |
0.0700924547 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_056691_060 |
0.0705179126 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009486_180 |
0.0714148104 |
- |
- |
unknown [Glycine max] |
| 16 |
Hb_003734_010 |
0.072521575 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 17 |
Hb_002631_170 |
0.0729128808 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 18 |
Hb_000603_030 |
0.0753496186 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 19 |
Hb_005402_040 |
0.0756908142 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 20 |
Hb_003645_060 |
0.0757100829 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |