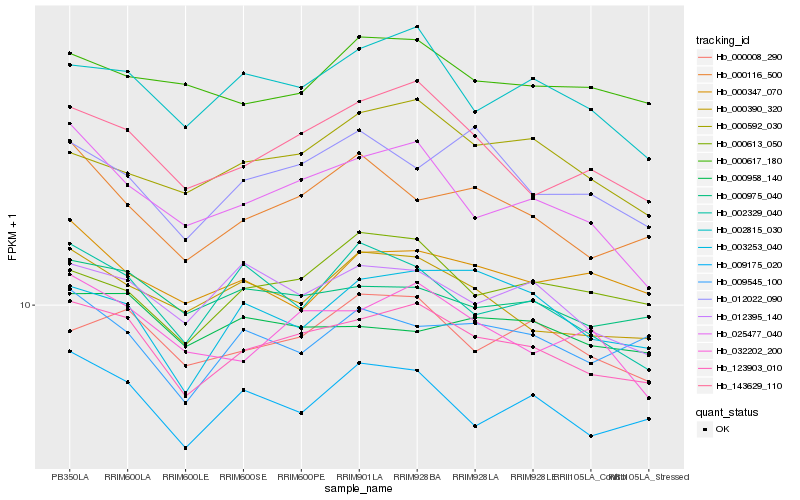
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123903_010 |
0.0 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_143629_110 |
0.0582921418 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 3 |
Hb_009175_020 |
0.0602956524 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_002815_030 |
0.0620542616 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 5 |
Hb_000975_040 |
0.0630263116 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 6 |
Hb_000390_320 |
0.0632458211 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 7 |
Hb_032202_200 |
0.063984752 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000008_290 |
0.064978971 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 9 |
Hb_012395_140 |
0.0654799934 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 10 |
Hb_025477_040 |
0.0664625701 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 11 |
Hb_000116_500 |
0.0670413962 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 12 |
Hb_003253_040 |
0.0683658142 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 13 |
Hb_000613_050 |
0.069802702 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 14 |
Hb_009545_100 |
0.0699368279 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 15 |
Hb_002329_040 |
0.0739589665 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 16 |
Hb_000958_140 |
0.0742975447 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 17 |
Hb_012022_090 |
0.0743435722 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Gossypium raimondii] |
| 18 |
Hb_000617_180 |
0.0752031604 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 19 |
Hb_000347_070 |
0.0754588853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000592_030 |
0.0762767401 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |