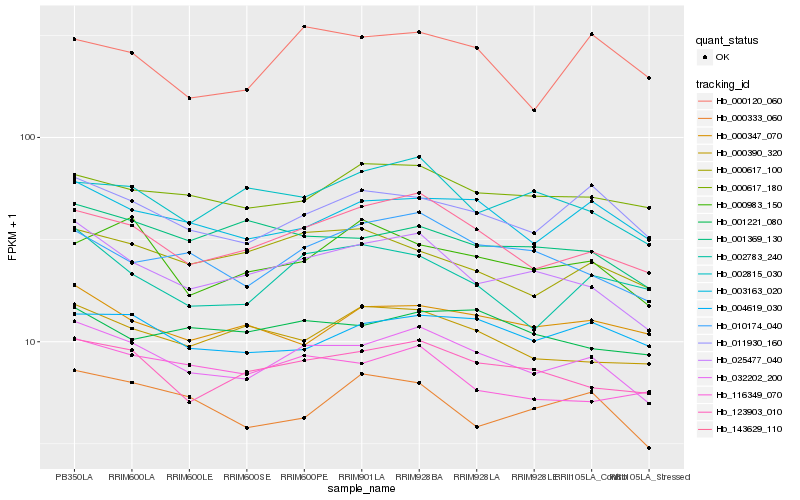
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_200 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_143629_110 |
0.0571348934 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 3 |
Hb_025477_040 |
0.0607566328 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 4 |
Hb_123903_010 |
0.063984752 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003163_020 |
0.0676438971 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_011930_160 |
0.0684381374 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 38-like [Jatropha curcas] |
| 7 |
Hb_010174_040 |
0.0765560738 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 8 |
Hb_004619_030 |
0.0803502551 |
- |
- |
PREDICTED: uncharacterized protein LOC105633364 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000617_180 |
0.0810026229 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 10 |
Hb_000390_320 |
0.0818315525 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 11 |
Hb_001369_130 |
0.0829932807 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000617_100 |
0.085069542 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 13 |
Hb_001221_080 |
0.0851795992 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_116349_070 |
0.0852602255 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 15 |
Hb_002783_240 |
0.0857499712 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 16 |
Hb_000347_070 |
0.0868054644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002815_030 |
0.0868773752 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 18 |
Hb_000120_060 |
0.0886102803 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 19 |
Hb_000333_060 |
0.0889470425 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 20 |
Hb_000983_150 |
0.0899871335 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |